Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Scrt2
Z-value: 0.88
Transcription factors associated with Scrt2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Scrt2
|
ENSMUSG00000060257.3 | Scrt2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Scrt2 | mm39_v1_chr2_+_151923449_151923544 | -0.01 | 9.3e-01 | Click! |
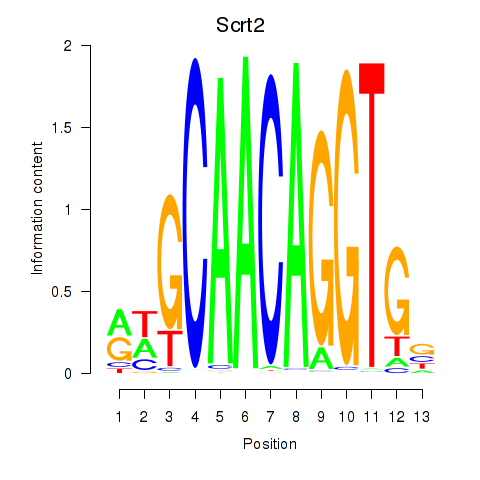
Activity profile of Scrt2 motif
Sorted Z-values of Scrt2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Scrt2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_36598263 | 3.63 |
ENSMUST00000155116.2
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr5_-_137145030 | 2.19 |
ENSMUST00000054384.6
ENSMUST00000152207.2 |
Trim56
|
tripartite motif-containing 56 |
| chr7_+_126690525 | 2.14 |
ENSMUST00000056288.7
ENSMUST00000206102.2 |
AI467606
|
expressed sequence AI467606 |
| chr6_-_86646118 | 2.03 |
ENSMUST00000001184.10
|
Mxd1
|
MAX dimerization protein 1 |
| chrX_+_149330371 | 2.01 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr4_+_98812082 | 1.99 |
ENSMUST00000091358.11
|
Usp1
|
ubiquitin specific peptidase 1 |
| chr2_-_30720345 | 1.86 |
ENSMUST00000041726.4
|
Asb6
|
ankyrin repeat and SOCS box-containing 6 |
| chr4_+_98812047 | 1.81 |
ENSMUST00000030289.9
|
Usp1
|
ubiquitin specific peptidase 1 |
| chr12_-_40087393 | 1.74 |
ENSMUST00000146905.2
|
Arl4a
|
ADP-ribosylation factor-like 4A |
| chr7_+_89779564 | 1.61 |
ENSMUST00000208742.2
ENSMUST00000049537.9 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr3_-_89009153 | 1.57 |
ENSMUST00000199668.3
ENSMUST00000196709.5 |
Fdps
|
farnesyl diphosphate synthetase |
| chr15_+_44320918 | 1.55 |
ENSMUST00000038336.12
ENSMUST00000166957.2 ENSMUST00000209244.2 |
Pkhd1l1
|
polycystic kidney and hepatic disease 1-like 1 |
| chr13_+_23940964 | 1.54 |
ENSMUST00000102965.4
|
H4c2
|
H4 clustered histone 2 |
| chr3_-_89009214 | 1.52 |
ENSMUST00000081848.13
|
Fdps
|
farnesyl diphosphate synthetase |
| chr18_+_60880149 | 1.48 |
ENSMUST00000236652.2
ENSMUST00000235966.2 |
Rps14
|
ribosomal protein S14 |
| chr7_+_89779421 | 1.47 |
ENSMUST00000207225.2
ENSMUST00000207484.2 ENSMUST00000209068.2 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr9_+_59563872 | 1.39 |
ENSMUST00000215623.2
ENSMUST00000215660.2 ENSMUST00000217353.2 |
Pkm
|
pyruvate kinase, muscle |
| chr5_-_138169509 | 1.36 |
ENSMUST00000153867.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr9_+_59563838 | 1.34 |
ENSMUST00000163694.4
|
Pkm
|
pyruvate kinase, muscle |
| chrX_-_48877080 | 1.30 |
ENSMUST00000114893.8
|
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr7_+_89779493 | 1.18 |
ENSMUST00000208730.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chrX_+_73473277 | 1.13 |
ENSMUST00000114127.8
ENSMUST00000064407.10 ENSMUST00000156707.3 |
Ikbkg
|
inhibitor of kappaB kinase gamma |
| chr3_-_107993906 | 1.12 |
ENSMUST00000102638.8
ENSMUST00000102637.8 |
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr5_-_138169476 | 0.98 |
ENSMUST00000147920.2
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr6_-_120334382 | 0.96 |
ENSMUST00000032283.12
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr1_-_192880260 | 0.91 |
ENSMUST00000161367.2
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr1_+_183078851 | 0.90 |
ENSMUST00000193625.2
|
Aida
|
axin interactor, dorsalization associated |
| chr7_-_100504610 | 0.89 |
ENSMUST00000156855.8
|
Relt
|
RELT tumor necrosis factor receptor |
| chr5_+_66833434 | 0.89 |
ENSMUST00000031131.11
|
Uchl1
|
ubiquitin carboxy-terminal hydrolase L1 |
| chr11_+_80320558 | 0.86 |
ENSMUST00000173565.2
|
Psmd11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr2_-_60711706 | 0.85 |
ENSMUST00000164147.8
ENSMUST00000112509.2 |
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr7_+_24990596 | 0.80 |
ENSMUST00000164820.2
|
Cic
|
capicua transcriptional repressor |
| chr7_-_127805518 | 0.74 |
ENSMUST00000033049.9
|
Cox6a2
|
cytochrome c oxidase subunit 6A2 |
| chr15_+_34306812 | 0.74 |
ENSMUST00000226766.2
ENSMUST00000163455.9 ENSMUST00000022947.7 ENSMUST00000228570.2 ENSMUST00000227759.2 |
Matn2
|
matrilin 2 |
| chr6_-_120334400 | 0.73 |
ENSMUST00000112703.8
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr8_-_103512274 | 0.70 |
ENSMUST00000075190.5
|
Cdh11
|
cadherin 11 |
| chr16_+_44167484 | 0.70 |
ENSMUST00000050897.7
|
Spice1
|
spindle and centriole associated protein 1 |
| chr5_-_38718967 | 0.69 |
ENSMUST00000201260.4
|
Wdr1
|
WD repeat domain 1 |
| chr18_-_70186503 | 0.69 |
ENSMUST00000117692.8
ENSMUST00000069749.9 |
Rab27b
|
RAB27B, member RAS oncogene family |
| chr2_+_31840151 | 0.65 |
ENSMUST00000001920.13
ENSMUST00000151276.3 |
Aif1l
|
allograft inflammatory factor 1-like |
| chr4_-_45084564 | 0.65 |
ENSMUST00000052236.13
|
Fbxo10
|
F-box protein 10 |
| chr3_-_53564759 | 0.63 |
ENSMUST00000091137.6
|
Frem2
|
Fras1 related extracellular matrix protein 2 |
| chr2_+_112097087 | 0.63 |
ENSMUST00000110987.9
ENSMUST00000028549.14 |
Slc12a6
|
solute carrier family 12, member 6 |
| chr2_-_169973076 | 0.63 |
ENSMUST00000063710.13
|
Zfp217
|
zinc finger protein 217 |
| chr1_-_133352115 | 0.61 |
ENSMUST00000153799.8
|
Sox13
|
SRY (sex determining region Y)-box 13 |
| chr4_+_129083553 | 0.60 |
ENSMUST00000106054.4
|
Yars
|
tyrosyl-tRNA synthetase |
| chr7_+_126446588 | 0.59 |
ENSMUST00000141805.8
ENSMUST00000064110.14 ENSMUST00000205938.2 ENSMUST00000152051.8 |
Doc2a
|
double C2, alpha |
| chr12_+_4642987 | 0.58 |
ENSMUST00000062580.8
ENSMUST00000220311.2 |
Itsn2
|
intersectin 2 |
| chr10_-_19727165 | 0.57 |
ENSMUST00000059805.6
|
Slc35d3
|
solute carrier family 35, member D3 |
| chr3_-_20209260 | 0.56 |
ENSMUST00000178328.8
|
Gyg
|
glycogenin |
| chr2_+_112096154 | 0.56 |
ENSMUST00000110991.9
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr3_-_20209220 | 0.56 |
ENSMUST00000184552.2
|
Gyg
|
glycogenin |
| chr5_-_138170077 | 0.56 |
ENSMUST00000155902.8
ENSMUST00000148879.8 |
Mcm7
|
minichromosome maintenance complex component 7 |
| chr17_-_56019571 | 0.56 |
ENSMUST00000086876.7
ENSMUST00000233557.2 ENSMUST00000233972.2 |
Pot1b
|
protection of telomeres 1B |
| chr7_-_119078472 | 0.55 |
ENSMUST00000209095.2
ENSMUST00000033263.6 ENSMUST00000207261.2 |
Umod
|
uromodulin |
| chr3_+_129974531 | 0.54 |
ENSMUST00000080335.11
ENSMUST00000106353.2 |
Col25a1
|
collagen, type XXV, alpha 1 |
| chr9_-_50650663 | 0.54 |
ENSMUST00000117093.2
ENSMUST00000121634.8 |
Dixdc1
|
DIX domain containing 1 |
| chr3_-_108747792 | 0.53 |
ENSMUST00000106596.4
ENSMUST00000102621.11 ENSMUST00000138552.6 |
Stxbp3
|
syntaxin binding protein 3 |
| chr4_-_135221926 | 0.52 |
ENSMUST00000102549.10
|
Nipal3
|
NIPA-like domain containing 3 |
| chr5_+_137627431 | 0.51 |
ENSMUST00000176667.8
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr13_+_109397184 | 0.48 |
ENSMUST00000153234.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr5_-_38719025 | 0.47 |
ENSMUST00000005234.13
|
Wdr1
|
WD repeat domain 1 |
| chr6_-_120334302 | 0.45 |
ENSMUST00000163827.8
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr12_+_65083093 | 0.43 |
ENSMUST00000120580.8
|
Prpf39
|
pre-mRNA processing factor 39 |
| chr8_+_31677345 | 0.42 |
ENSMUST00000066173.12
ENSMUST00000161788.8 ENSMUST00000110527.9 ENSMUST00000161502.2 |
Fut10
|
fucosyltransferase 10 |
| chr15_-_11907267 | 0.40 |
ENSMUST00000228489.2
|
Npr3
|
natriuretic peptide receptor 3 |
| chr4_-_129083335 | 0.39 |
ENSMUST00000106061.9
ENSMUST00000072431.13 |
S100pbp
|
S100P binding protein |
| chr7_+_44465353 | 0.38 |
ENSMUST00000208626.2
ENSMUST00000057195.17 |
Nup62
|
nucleoporin 62 |
| chr15_-_66432938 | 0.38 |
ENSMUST00000048372.7
|
Tmem71
|
transmembrane protein 71 |
| chr8_-_4309257 | 0.38 |
ENSMUST00000053252.9
|
Ctxn1
|
cortexin 1 |
| chr7_-_119078330 | 0.38 |
ENSMUST00000207460.2
|
Umod
|
uromodulin |
| chr11_+_71640739 | 0.36 |
ENSMUST00000150531.2
|
Wscd1
|
WSC domain containing 1 |
| chr7_+_122977205 | 0.36 |
ENSMUST00000033025.7
ENSMUST00000206574.2 |
Lcmt1
|
leucine carboxyl methyltransferase 1 |
| chr7_-_45480200 | 0.36 |
ENSMUST00000107723.9
ENSMUST00000131384.3 |
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chr6_-_57885013 | 0.35 |
ENSMUST00000228076.2
ENSMUST00000228905.2 ENSMUST00000227650.2 ENSMUST00000227342.2 ENSMUST00000228257.2 |
Vmn1r22
|
vomeronasal 1 receptor 22 |
| chrX_-_52203405 | 0.35 |
ENSMUST00000114843.9
|
Plac1
|
placental specific protein 1 |
| chr5_-_123822351 | 0.34 |
ENSMUST00000111564.8
ENSMUST00000063905.12 |
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr7_+_127603083 | 0.34 |
ENSMUST00000106248.8
|
Trim72
|
tripartite motif-containing 72 |
| chr11_+_87483723 | 0.34 |
ENSMUST00000119628.8
|
Mtmr4
|
myotubularin related protein 4 |
| chr4_+_98812144 | 0.33 |
ENSMUST00000169053.2
|
Usp1
|
ubiquitin specific peptidase 1 |
| chr7_-_29898236 | 0.33 |
ENSMUST00000001845.13
|
Capns1
|
calpain, small subunit 1 |
| chr12_+_78795763 | 0.32 |
ENSMUST00000082024.7
|
Mpp5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
| chr4_+_124960465 | 0.31 |
ENSMUST00000052183.7
|
Snip1
|
Smad nuclear interacting protein 1 |
| chr16_+_31998588 | 0.31 |
ENSMUST00000231941.2
|
Bex6
|
brain expressed family member 6 |
| chr4_-_41731142 | 0.30 |
ENSMUST00000171251.8
|
Arid3c
|
AT rich interactive domain 3C (BRIGHT-like) |
| chr14_+_55120875 | 0.29 |
ENSMUST00000134077.2
ENSMUST00000172844.8 ENSMUST00000133397.4 ENSMUST00000227108.2 |
Gm20521
Bcl2l2
|
predicted gene 20521 BCL2-like 2 |
| chr3_-_122778052 | 0.29 |
ENSMUST00000199401.2
ENSMUST00000197314.5 ENSMUST00000197934.5 ENSMUST00000090379.7 |
Usp53
|
ubiquitin specific peptidase 53 |
| chr2_+_152753231 | 0.28 |
ENSMUST00000028970.8
|
Mylk2
|
myosin, light polypeptide kinase 2, skeletal muscle |
| chr10_+_79852487 | 0.28 |
ENSMUST00000099501.10
|
Arhgap45
|
Rho GTPase activating protein 45 |
| chr17_+_44499451 | 0.27 |
ENSMUST00000024755.7
|
Clic5
|
chloride intracellular channel 5 |
| chr2_-_53975501 | 0.27 |
ENSMUST00000100089.3
|
Rprm
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr5_-_123822338 | 0.27 |
ENSMUST00000111561.8
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr3_-_108747767 | 0.26 |
ENSMUST00000196679.5
|
Stxbp3
|
syntaxin binding protein 3 |
| chr12_-_115206715 | 0.25 |
ENSMUST00000103527.2
ENSMUST00000194071.2 |
Ighv1-56
|
immunoglobulin heavy variable 1-56 |
| chr1_-_156766957 | 0.24 |
ENSMUST00000171292.8
ENSMUST00000063199.13 ENSMUST00000027886.14 |
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr9_-_78015866 | 0.23 |
ENSMUST00000162625.2
ENSMUST00000159099.8 ENSMUST00000085311.13 |
Fbxo9
|
f-box protein 9 |
| chr1_-_156767123 | 0.22 |
ENSMUST00000189316.7
ENSMUST00000190648.7 ENSMUST00000172057.8 ENSMUST00000191605.7 |
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr2_+_31840340 | 0.21 |
ENSMUST00000148056.4
|
Aif1l
|
allograft inflammatory factor 1-like |
| chr6_-_148732946 | 0.20 |
ENSMUST00000048418.14
|
Ipo8
|
importin 8 |
| chr2_+_121188195 | 0.19 |
ENSMUST00000125812.8
ENSMUST00000078222.9 ENSMUST00000125221.3 ENSMUST00000150271.8 |
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous |
| chr8_-_85663976 | 0.19 |
ENSMUST00000109741.9
ENSMUST00000119820.2 |
Mast1
|
microtubule associated serine/threonine kinase 1 |
| chrX_-_100128958 | 0.18 |
ENSMUST00000101362.8
ENSMUST00000073927.5 |
Slc7a3
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3 |
| chr19_+_47167554 | 0.18 |
ENSMUST00000235290.2
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr9_-_27066685 | 0.16 |
ENSMUST00000034472.16
|
Jam3
|
junction adhesion molecule 3 |
| chr7_+_100742070 | 0.16 |
ENSMUST00000208439.2
|
Fchsd2
|
FCH and double SH3 domains 2 |
| chr5_+_89675288 | 0.15 |
ENSMUST00000048557.3
|
Npffr2
|
neuropeptide FF receptor 2 |
| chr14_-_61495832 | 0.15 |
ENSMUST00000121148.8
|
Sgcg
|
sarcoglycan, gamma (dystrophin-associated glycoprotein) |
| chr14_-_61495934 | 0.15 |
ENSMUST00000077954.13
|
Sgcg
|
sarcoglycan, gamma (dystrophin-associated glycoprotein) |
| chr12_+_78795647 | 0.13 |
ENSMUST00000219667.2
|
Mpp5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
| chr4_-_135221810 | 0.13 |
ENSMUST00000105856.9
|
Nipal3
|
NIPA-like domain containing 3 |
| chr3_+_37402795 | 0.13 |
ENSMUST00000200585.5
ENSMUST00000038885.10 |
Fgf2
|
fibroblast growth factor 2 |
| chr3_-_124219673 | 0.13 |
ENSMUST00000029598.10
|
1700006A11Rik
|
RIKEN cDNA 1700006A11 gene |
| chr18_-_82424902 | 0.13 |
ENSMUST00000065224.8
|
Galr1
|
galanin receptor 1 |
| chr11_+_77384234 | 0.12 |
ENSMUST00000037285.10
ENSMUST00000100812.4 |
Git1
|
GIT ArfGAP 1 |
| chr12_-_11200306 | 0.12 |
ENSMUST00000055673.2
|
Kcns3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| chr15_+_102898966 | 0.11 |
ENSMUST00000001703.8
|
Hoxc8
|
homeobox C8 |
| chr4_-_151129020 | 0.11 |
ENSMUST00000103204.11
|
Per3
|
period circadian clock 3 |
| chr4_-_155947819 | 0.10 |
ENSMUST00000030949.4
|
Tas1r3
|
taste receptor, type 1, member 3 |
| chr13_-_98342659 | 0.10 |
ENSMUST00000109426.3
|
Arhgef28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr1_-_183150867 | 0.09 |
ENSMUST00000194543.4
|
Mia3
|
melanoma inhibitory activity 3 |
| chr15_-_97902515 | 0.09 |
ENSMUST00000088355.12
|
Col2a1
|
collagen, type II, alpha 1 |
| chr11_+_115225557 | 0.08 |
ENSMUST00000106543.8
ENSMUST00000019006.5 |
Otop3
|
otopetrin 3 |
| chr4_-_129083392 | 0.08 |
ENSMUST00000117497.8
ENSMUST00000117350.2 |
S100pbp
|
S100P binding protein |
| chr6_+_78382131 | 0.08 |
ENSMUST00000023906.4
|
Reg2
|
regenerating islet-derived 2 |
| chr4_-_129083439 | 0.08 |
ENSMUST00000106059.8
|
S100pbp
|
S100P binding protein |
| chr7_-_130931235 | 0.07 |
ENSMUST00000188899.2
|
Fam24b
|
family with sequence similarity 24 member B |
| chr7_-_24211879 | 0.07 |
ENSMUST00000234781.2
|
Gm50092
|
predicted gene, 50092 |
| chr10_+_116013122 | 0.07 |
ENSMUST00000148731.8
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr1_-_156766381 | 0.06 |
ENSMUST00000188656.7
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr11_-_76514334 | 0.06 |
ENSMUST00000238684.2
|
Abr
|
active BCR-related gene |
| chr19_-_61216834 | 0.06 |
ENSMUST00000076046.7
|
Csf2ra
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr10_+_116013256 | 0.06 |
ENSMUST00000155606.8
ENSMUST00000128399.2 |
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr7_+_126447080 | 0.05 |
ENSMUST00000147257.2
ENSMUST00000139174.2 |
Doc2a
|
double C2, alpha |
| chr1_+_182236728 | 0.05 |
ENSMUST00000117245.2
|
Trp53bp2
|
transformation related protein 53 binding protein 2 |
| chr17_+_34573760 | 0.05 |
ENSMUST00000178562.2
ENSMUST00000025198.15 |
Btnl2
|
butyrophilin-like 2 |
| chr8_+_47192911 | 0.05 |
ENSMUST00000208433.2
|
Irf2
|
interferon regulatory factor 2 |
| chr10_-_79973210 | 0.05 |
ENSMUST00000170219.9
ENSMUST00000169546.9 |
Cbarp
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein |
| chr11_-_107805830 | 0.04 |
ENSMUST00000039071.3
|
Cacng5
|
calcium channel, voltage-dependent, gamma subunit 5 |
| chr12_-_17374704 | 0.04 |
ENSMUST00000020884.16
ENSMUST00000095820.12 ENSMUST00000221129.2 ENSMUST00000127185.8 |
Atp6v1c2
|
ATPase, H+ transporting, lysosomal V1 subunit C2 |
| chr1_-_156766351 | 0.04 |
ENSMUST00000189648.2
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr10_-_109669053 | 0.03 |
ENSMUST00000238286.2
|
Nav3
|
neuron navigator 3 |
| chr15_-_71599664 | 0.03 |
ENSMUST00000022953.10
|
Fam135b
|
family with sequence similarity 135, member B |
| chr11_+_110956980 | 0.03 |
ENSMUST00000042970.3
|
Kcnj2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chrX_-_71318353 | 0.03 |
ENSMUST00000064780.4
|
Gabre
|
gamma-aminobutyric acid (GABA) A receptor, subunit epsilon |
| chr10_+_56253418 | 0.02 |
ENSMUST00000068581.9
ENSMUST00000217789.2 |
Gja1
|
gap junction protein, alpha 1 |
| chr6_-_53797748 | 0.02 |
ENSMUST00000127748.5
|
Tril
|
TLR4 interactor with leucine-rich repeats |
| chr4_-_133739697 | 0.02 |
ENSMUST00000176113.8
|
Lin28a
|
lin-28 homolog A (C. elegans) |
| chr15_-_78602971 | 0.02 |
ENSMUST00000088592.6
|
Elfn2
|
leucine rich repeat and fibronectin type III, extracellular 2 |
| chr14_+_9646630 | 0.01 |
ENSMUST00000112658.8
ENSMUST00000112657.9 ENSMUST00000177814.2 ENSMUST00000067491.14 |
Cadps
|
Ca2+-dependent secretion activator |
| chr15_-_77331660 | 0.00 |
ENSMUST00000089469.7
|
Apol7b
|
apolipoprotein L 7b |
| chr2_-_85997866 | 0.00 |
ENSMUST00000213949.2
ENSMUST00000213886.2 ENSMUST00000216028.2 |
Olfr1043
|
olfactory receptor 1043 |
| chr16_+_42727926 | 0.00 |
ENSMUST00000151244.8
ENSMUST00000114694.9 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.7 | 4.3 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.5 | 3.6 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.3 | 4.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.3 | 1.2 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.3 | 2.7 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.2 | 0.7 | GO:0031104 | dendrite regeneration(GO:0031104) |
| 0.2 | 2.9 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 0.9 | GO:0072023 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.2 | 0.9 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.2 | 1.2 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 0.6 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.2 | 0.6 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.1 | 0.9 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.6 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 1.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 2.0 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 2.2 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 0.4 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 0.3 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.1 | 0.5 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 0.5 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.1 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.1 | 0.4 | GO:1903436 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.1 | 0.3 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.1 | 0.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.5 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 1.1 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.4 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.5 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.6 | GO:0022615 | protein to membrane docking(GO:0022615) |
| 0.0 | 0.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.5 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.7 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.0 | 0.7 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0015819 | lysine transport(GO:0015819) |
| 0.0 | 0.5 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.7 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.6 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 | 0.1 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 0.0 | 0.3 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.0 | 0.5 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 1.7 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 1.1 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.4 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.2 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.2 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.6 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.6 | 4.3 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.4 | 1.2 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.2 | 2.9 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 3.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.8 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.1 | 0.7 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 1.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.9 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.5 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 0.6 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.6 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.6 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.4 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 1.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 4.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.0 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.7 | 2.0 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.5 | 2.7 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.3 | 1.2 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.3 | 4.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 0.9 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.1 | 1.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 1.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 3.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 1.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.4 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.1 | 1.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 0.9 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 0.4 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.1 | 2.9 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 0.4 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 0.6 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.6 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.4 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.2 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 0.0 | 0.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 1.1 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 4.1 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.2 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.9 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.6 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.1 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 4.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 2.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.2 | 2.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 3.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 3.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 2.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 4.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.9 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.5 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |