Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
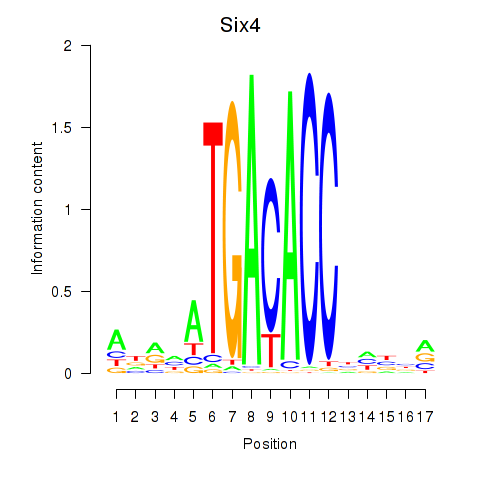
Results for Six4
Z-value: 0.42
Transcription factors associated with Six4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Six4
|
ENSMUSG00000034460.10 | Six4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Six4 | mm39_v1_chr12_-_73160181_73160230 | 0.01 | 9.7e-01 | Click! |
Activity profile of Six4 motif
Sorted Z-values of Six4 motif
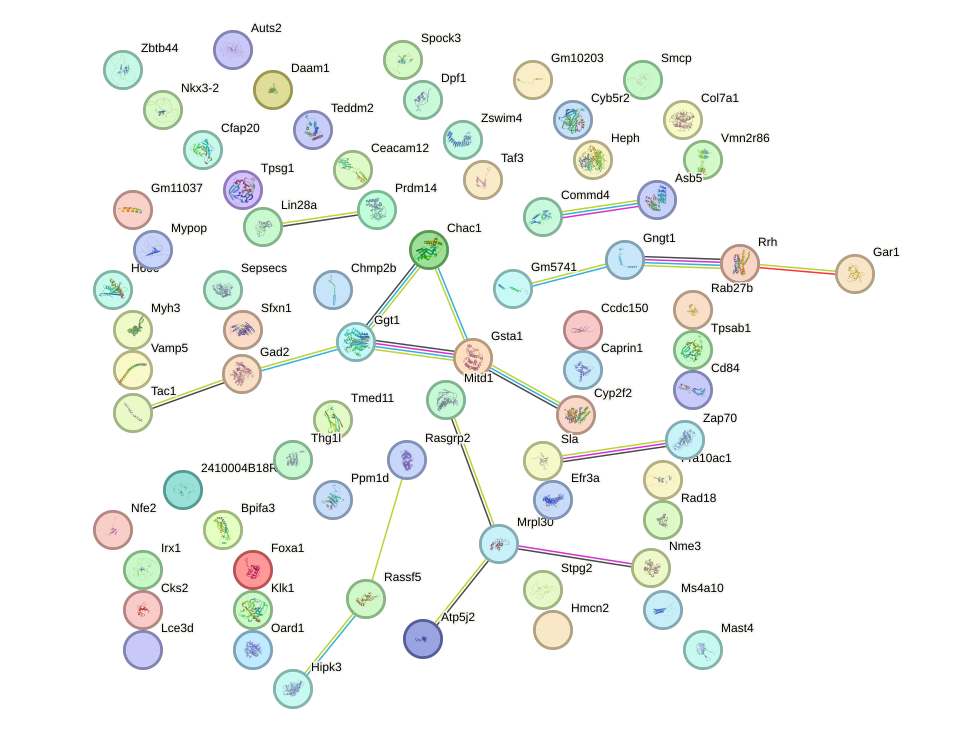
Network of associatons between targets according to the STRING database.
First level regulatory network of Six4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_78137927 | 0.99 |
ENSMUST00000098537.4
|
Gsta1
|
glutathione S-transferase, alpha 1 (Ya) |
| chr9_+_78197205 | 0.69 |
ENSMUST00000119823.8
ENSMUST00000121273.2 |
Gsta5
|
glutathione S-transferase alpha 5 |
| chr15_-_103163860 | 0.43 |
ENSMUST00000075192.13
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr10_+_75399920 | 0.32 |
ENSMUST00000141062.8
ENSMUST00000152657.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr15_+_65658874 | 0.32 |
ENSMUST00000015146.16
ENSMUST00000173858.8 ENSMUST00000172756.2 ENSMUST00000174856.7 |
Efr3a
|
EFR3 homolog A |
| chr17_+_48717007 | 0.30 |
ENSMUST00000167180.8
ENSMUST00000046651.7 ENSMUST00000233426.2 |
Oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr3_+_40905216 | 0.29 |
ENSMUST00000191872.6
ENSMUST00000200432.2 |
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr1_-_37929414 | 0.28 |
ENSMUST00000139725.8
ENSMUST00000027257.10 |
Mitd1
|
MIT, microtubule interacting and transport, domain containing 1 |
| chr3_+_40905066 | 0.28 |
ENSMUST00000191805.7
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr16_-_65359406 | 0.27 |
ENSMUST00000231259.2
|
Chmp2b
|
charged multivesicular body protein 2B |
| chr19_-_38212544 | 0.26 |
ENSMUST00000067167.6
|
Fra10ac1
|
FRA10AC1 homolog (human) |
| chr2_+_119181703 | 0.25 |
ENSMUST00000028780.4
|
Chac1
|
ChaC, cation transport regulator 1 |
| chr11_-_45845992 | 0.25 |
ENSMUST00000109254.2
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr2_-_103627937 | 0.24 |
ENSMUST00000028607.13
|
Caprin1
|
cell cycle associated protein 1 |
| chr19_+_6450641 | 0.23 |
ENSMUST00000113467.2
|
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr16_-_65359566 | 0.22 |
ENSMUST00000004965.8
|
Chmp2b
|
charged multivesicular body protein 2B |
| chrX_-_149223883 | 0.21 |
ENSMUST00000163233.2
|
Tmem29
|
transmembrane protein 29 |
| chr3_-_92493507 | 0.20 |
ENSMUST00000194965.6
|
Smcp
|
sperm mitochondria-associated cysteine-rich protein |
| chr13_-_72111832 | 0.19 |
ENSMUST00000077337.9
|
Irx1
|
Iroquois homeobox 1 |
| chr13_+_54225828 | 0.19 |
ENSMUST00000021930.10
|
Sfxn1
|
sideroflexin 1 |
| chr3_-_75864195 | 0.18 |
ENSMUST00000038563.14
ENSMUST00000167078.8 ENSMUST00000117242.8 |
Golim4
|
golgi integral membrane protein 4 |
| chr12_+_71936500 | 0.18 |
ENSMUST00000221317.2
|
Daam1
|
dishevelled associated activator of morphogenesis 1 |
| chr7_+_18725170 | 0.18 |
ENSMUST00000059331.9
ENSMUST00000131087.2 |
Mypop
|
Myb-related transcription factor, partner of profilin |
| chr18_-_70186503 | 0.17 |
ENSMUST00000117692.8
ENSMUST00000069749.9 |
Rab27b
|
RAB27B, member RAS oncogene family |
| chr1_+_37929558 | 0.17 |
ENSMUST00000027256.12
ENSMUST00000193673.6 ENSMUST00000160082.3 |
Mrpl30
|
mitochondrial ribosomal protein L30 |
| chr2_-_10053381 | 0.17 |
ENSMUST00000026888.11
|
Taf3
|
TATA-box binding protein associated factor 3 |
| chr4_-_133746138 | 0.16 |
ENSMUST00000051674.3
|
Lin28a
|
lin-28 homolog A (C. elegans) |
| chr11_-_45846291 | 0.16 |
ENSMUST00000011398.13
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr7_+_26819334 | 0.14 |
ENSMUST00000003100.10
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2 |
| chr17_-_25564501 | 0.14 |
ENSMUST00000153118.2
ENSMUST00000146856.3 |
Tpsab1
|
tryptase alpha/beta 1 |
| chr7_+_27879650 | 0.14 |
ENSMUST00000172467.8
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr19_-_6957789 | 0.13 |
ENSMUST00000070878.9
ENSMUST00000177752.9 |
Fkbp2
|
FK506 binding protein 2 |
| chr5_-_132570710 | 0.13 |
ENSMUST00000182974.9
|
Auts2
|
autism susceptibility candidate 2 |
| chr5_-_41921834 | 0.12 |
ENSMUST00000060820.8
|
Nkx3-2
|
NK3 homeobox 2 |
| chr12_-_57592907 | 0.11 |
ENSMUST00000044380.8
|
Foxa1
|
forkhead box A1 |
| chr19_+_6450553 | 0.10 |
ENSMUST00000146831.8
ENSMUST00000035716.15 ENSMUST00000138555.8 ENSMUST00000167240.8 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr16_+_11223512 | 0.10 |
ENSMUST00000096273.9
|
Snx29
|
sorting nexin 29 |
| chr17_+_25588241 | 0.10 |
ENSMUST00000160377.8
ENSMUST00000160485.8 |
Tpsg1
|
tryptase gamma 1 |
| chr6_-_112673565 | 0.10 |
ENSMUST00000113182.8
ENSMUST00000113180.8 ENSMUST00000068487.12 ENSMUST00000077088.11 |
Rad18
|
RAD18 E3 ubiquitin protein ligase |
| chr1_-_131127825 | 0.10 |
ENSMUST00000068564.15
|
Rassf5
|
Ras association (RalGDS/AF-6) domain family member 5 |
| chr9_+_30941924 | 0.09 |
ENSMUST00000216649.2
ENSMUST00000115222.10 |
Zbtb44
|
zinc finger and BTB domain containing 44 |
| chr5_-_145128321 | 0.09 |
ENSMUST00000161845.2
|
Atp5j2
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F2 |
| chr7_+_17799889 | 0.09 |
ENSMUST00000108483.2
|
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chr1_+_171667259 | 0.09 |
ENSMUST00000155802.9
|
Cd84
|
CD84 antigen |
| chr8_+_55024446 | 0.09 |
ENSMUST00000239166.2
ENSMUST00000239106.2 ENSMUST00000239152.2 |
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr1_+_36806015 | 0.08 |
ENSMUST00000185871.2
|
Zap70
|
zeta-chain (TCR) associated protein kinase |
| chr9_-_57065572 | 0.08 |
ENSMUST00000065358.9
|
Commd4
|
COMM domain containing 4 |
| chr7_+_17799849 | 0.08 |
ENSMUST00000032520.9
|
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chr7_+_17799863 | 0.08 |
ENSMUST00000108487.9
|
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chr9_+_108782664 | 0.07 |
ENSMUST00000026740.6
|
Col7a1
|
collagen, type VII, alpha 1 |
| chr2_+_130975417 | 0.07 |
ENSMUST00000110225.2
|
Gm11037
|
predicted gene 11037 |
| chr9_+_108782646 | 0.07 |
ENSMUST00000112070.8
|
Col7a1
|
collagen, type VII, alpha 1 |
| chr11_-_34048538 | 0.07 |
ENSMUST00000223852.2
|
4930469K13Rik
|
RIKEN cDNA 4930469K13 gene |
| chr8_-_85794611 | 0.07 |
ENSMUST00000177563.2
|
Gm5741
|
predicted gene 5741 |
| chr7_+_43474819 | 0.07 |
ENSMUST00000107967.3
|
Klk6
|
kallikrein related-peptidase 6 |
| chr19_-_10952028 | 0.07 |
ENSMUST00000072748.13
|
Ms4a10
|
membrane-spanning 4-domains, subfamily A, member 10 |
| chrX_-_149224054 | 0.07 |
ENSMUST00000059256.8
|
Tmem29
|
transmembrane protein 29 |
| chr17_+_47908025 | 0.07 |
ENSMUST00000183206.2
|
Ccnd3
|
cyclin D3 |
| chr11_+_66969119 | 0.07 |
ENSMUST00000108689.8
ENSMUST00000007301.14 ENSMUST00000165221.2 |
Myh3
|
myosin, heavy polypeptide 3, skeletal muscle, embryonic |
| chrX_+_139243012 | 0.07 |
ENSMUST00000208130.2
|
Frmpd3
|
FERM and PDZ domain containing 3 |
| chr2_+_31204314 | 0.06 |
ENSMUST00000113532.9
|
Hmcn2
|
hemicentin 2 |
| chr6_+_3993774 | 0.06 |
ENSMUST00000031673.7
|
Gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr10_+_79590910 | 0.06 |
ENSMUST00000219981.2
ENSMUST00000219228.2 ENSMUST00000020577.4 |
Fgf22
|
fibroblast growth factor 22 |
| chr10_-_3217767 | 0.06 |
ENSMUST00000170893.3
|
H60c
|
histocompatibility 60c |
| chr10_-_3217718 | 0.06 |
ENSMUST00000216211.2
|
H60c
|
histocompatibility 60c |
| chr12_-_114160354 | 0.06 |
ENSMUST00000103476.2
ENSMUST00000191693.2 |
Ighv3-3
|
immunoglobulin heavy variable V3-3 |
| chr12_-_113726328 | 0.06 |
ENSMUST00000195381.2
|
Ighv5-12-4
|
immunoglobulin heavy variable 5-12-4 |
| chr8_+_63404395 | 0.06 |
ENSMUST00000119068.8
|
Spock3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 3 |
| chr5_-_108943211 | 0.05 |
ENSMUST00000004943.2
|
Tmed11
|
transmembrane p24 trafficking protein 11 |
| chr7_-_104542833 | 0.05 |
ENSMUST00000081116.2
|
Olfr666
|
olfactory receptor 666 |
| chr8_-_84963653 | 0.05 |
ENSMUST00000039480.7
|
Zswim4
|
zinc finger SWIM-type containing 4 |
| chr6_-_72357398 | 0.05 |
ENSMUST00000101285.10
ENSMUST00000074231.6 |
Vamp5
|
vesicle-associated membrane protein 5 |
| chr19_-_10952009 | 0.05 |
ENSMUST00000191343.7
|
Ms4a10
|
membrane-spanning 4-domains, subfamily A, member 10 |
| chr1_-_13197387 | 0.05 |
ENSMUST00000047577.7
|
Prdm14
|
PR domain containing 14 |
| chr13_-_103042554 | 0.04 |
ENSMUST00000171791.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr3_+_138058139 | 0.04 |
ENSMUST00000090166.5
|
Adh6b
|
alcohol dehydrogenase 6B (class V) |
| chr6_+_124908389 | 0.04 |
ENSMUST00000180095.4
|
Mlf2
|
myeloid leukemia factor 2 |
| chr1_-_153726930 | 0.04 |
ENSMUST00000059607.8
|
Teddm2
|
transmembrane epididymal family member 2 |
| chr19_+_29388312 | 0.04 |
ENSMUST00000112576.4
|
Pdcd1lg2
|
programmed cell death 1 ligand 2 |
| chr8_+_96429665 | 0.04 |
ENSMUST00000073139.14
ENSMUST00000080666.8 ENSMUST00000212160.2 |
Ndrg4
|
N-myc downstream regulated gene 4 |
| chr7_-_107357104 | 0.04 |
ENSMUST00000208217.2
ENSMUST00000052438.8 |
Cyb5r2
|
cytochrome b5 reductase 2 |
| chr2_+_153972253 | 0.04 |
ENSMUST00000028984.13
ENSMUST00000109746.8 |
Bpifa3
|
BPI fold containing family A, member 3 |
| chr9_+_19047613 | 0.04 |
ENSMUST00000215699.2
|
Olfr837
|
olfactory receptor 837 |
| chr6_+_124908439 | 0.03 |
ENSMUST00000032214.14
|
Mlf2
|
myeloid leukemia factor 2 |
| chr11_+_85202990 | 0.03 |
ENSMUST00000127717.2
|
Ppm1d
|
protein phosphatase 1D magnesium-dependent, delta isoform |
| chr16_-_20141815 | 0.03 |
ENSMUST00000023513.12
|
Cyp2ab1
|
cytochrome P450, family 2, subfamily ab, polypeptide 1 |
| chr6_+_149031668 | 0.03 |
ENSMUST00000087348.4
|
Gm10203
|
predicted gene 10203 |
| chr13_+_51799268 | 0.03 |
ENSMUST00000075853.6
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr4_-_144513121 | 0.03 |
ENSMUST00000105746.3
|
Aadacl4fm5
|
AADACL4 family member 5 |
| chr2_-_89951611 | 0.03 |
ENSMUST00000216493.2
ENSMUST00000214404.2 |
Olfr1269
|
olfactory receptor 1269 |
| chr2_-_104324035 | 0.03 |
ENSMUST00000111124.8
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr2_+_22512195 | 0.02 |
ENSMUST00000028123.4
|
Gad2
|
glutamic acid decarboxylase 2 |
| chr3_+_92864693 | 0.02 |
ENSMUST00000059053.11
|
Lce3d
|
late cornified envelope 3D |
| chr3_+_138911419 | 0.02 |
ENSMUST00000106239.8
|
Stpg2
|
sperm tail PG rich repeat containing 2 |
| chr3_+_138911648 | 0.02 |
ENSMUST00000062306.7
|
Stpg2
|
sperm tail PG rich repeat containing 2 |
| chr3_+_145643760 | 0.02 |
ENSMUST00000039571.14
|
2410004B18Rik
|
RIKEN cDNA 2410004B18 gene |
| chr3_-_129625023 | 0.02 |
ENSMUST00000029643.15
|
Gar1
|
GAR1 ribonucleoprotein |
| chr10_-_130291919 | 0.02 |
ENSMUST00000170257.4
|
Vmn2r86
|
vomeronasal 2, receptor 86 |
| chr17_+_25115461 | 0.02 |
ENSMUST00000024978.7
|
Nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr15_+_91949032 | 0.02 |
ENSMUST00000169825.8
|
Cntn1
|
contactin 1 |
| chr3_-_129616183 | 0.01 |
ENSMUST00000196902.5
|
Rrh
|
retinal pigment epithelium derived rhodopsin homolog |
| chr6_+_7555053 | 0.01 |
ENSMUST00000090679.9
ENSMUST00000184986.2 |
Tac1
|
tachykinin 1 |
| chr8_-_96161466 | 0.00 |
ENSMUST00000213086.2
ENSMUST00000034249.8 |
Cfap20
|
cilia and flagella associated protein 20 |
| chr1_+_54289833 | 0.00 |
ENSMUST00000027128.11
|
Ccdc150
|
coiled-coil domain containing 150 |
| chr6_+_124908341 | 0.00 |
ENSMUST00000203021.3
|
Mlf2
|
myeloid leukemia factor 2 |
| chr15_-_66703471 | 0.00 |
ENSMUST00000164163.8
|
Sla
|
src-like adaptor |
| chr7_+_29008840 | 0.00 |
ENSMUST00000137848.2
|
Dpf1
|
D4, zinc and double PHD fingers family 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:0090420 | naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 0.0 | 0.6 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.1 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.0 | 0.8 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 1.0 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 0.1 | GO:0032685 | negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) |
| 0.0 | 0.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.2 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.4 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 | 0.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0043366 | beta selection(GO:0043366) |
| 0.0 | 0.0 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.2 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 1.0 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |