Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Smad3
Z-value: 1.26
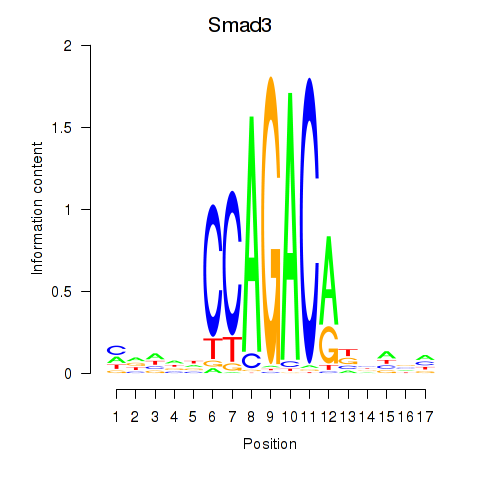
Transcription factors associated with Smad3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Smad3
|
ENSMUSG00000032402.13 | Smad3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Smad3 | mm39_v1_chr9_-_63665216_63665276 | -0.31 | 6.2e-02 | Click! |
Activity profile of Smad3 motif
Sorted Z-values of Smad3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Smad3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_60455331 | 3.93 |
ENSMUST00000135953.2
|
Mup1
|
major urinary protein 1 |
| chr13_+_23940964 | 2.35 |
ENSMUST00000102965.4
|
H4c2
|
H4 clustered histone 2 |
| chr13_+_22220000 | 2.17 |
ENSMUST00000110455.4
|
H2bc12
|
H2B clustered histone 12 |
| chr7_+_123061535 | 2.05 |
ENSMUST00000098056.6
|
Aqp8
|
aquaporin 8 |
| chr7_+_26534730 | 2.02 |
ENSMUST00000005685.15
|
Cyp2a5
|
cytochrome P450, family 2, subfamily a, polypeptide 5 |
| chr13_+_22227359 | 1.95 |
ENSMUST00000110452.2
|
H2bc11
|
H2B clustered histone 11 |
| chr7_+_26006594 | 1.83 |
ENSMUST00000098657.5
|
Cyp2a4
|
cytochrome P450, family 2, subfamily a, polypeptide 4 |
| chr13_-_23735822 | 1.80 |
ENSMUST00000102971.2
|
H4c6
|
H4 clustered histone 6 |
| chr13_+_23765546 | 1.64 |
ENSMUST00000102968.3
|
H4c4
|
H4 clustered histone 4 |
| chr13_-_22227114 | 1.46 |
ENSMUST00000091741.6
|
H2ac11
|
H2A clustered histone 11 |
| chr13_-_23755374 | 1.41 |
ENSMUST00000102969.6
|
H2ac8
|
H2A clustered histone 8 |
| chr13_+_23715220 | 1.39 |
ENSMUST00000102972.6
|
H4c8
|
H4 clustered histone 8 |
| chr13_+_23755551 | 1.34 |
ENSMUST00000079251.8
|
H2bc8
|
H2B clustered histone 8 |
| chr13_-_22017677 | 1.30 |
ENSMUST00000081342.7
|
H2ac24
|
H2A clustered histone 24 |
| chr13_+_23718038 | 1.28 |
ENSMUST00000073261.3
|
H2ac10
|
H2A clustered histone 10 |
| chr13_-_21934675 | 1.27 |
ENSMUST00000102983.2
|
H4c12
|
H4 clustered histone 12 |
| chr13_+_22017906 | 1.26 |
ENSMUST00000180288.2
|
H2bc24
|
H2B clustered histone 24 |
| chr15_-_103161237 | 1.23 |
ENSMUST00000154510.8
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr2_-_152673032 | 1.23 |
ENSMUST00000128172.3
|
Bcl2l1
|
BCL2-like 1 |
| chr2_-_152673585 | 1.22 |
ENSMUST00000156688.2
ENSMUST00000007803.12 |
Bcl2l1
|
BCL2-like 1 |
| chr13_+_21971631 | 1.10 |
ENSMUST00000110473.3
ENSMUST00000102982.2 |
H2bc22
|
H2B clustered histone 22 |
| chr2_+_29759495 | 1.10 |
ENSMUST00000047521.7
ENSMUST00000134152.2 |
Cercam
|
cerebral endothelial cell adhesion molecule |
| chr13_-_22016364 | 1.06 |
ENSMUST00000102979.2
|
H4c18
|
H4 clustered histone 18 |
| chr13_-_22225527 | 1.06 |
ENSMUST00000102977.4
|
H4c9
|
H4 clustered histone 9 |
| chr13_-_21900313 | 1.02 |
ENSMUST00000091756.2
|
H2bc13
|
H2B clustered histone 13 |
| chr6_+_136785230 | 0.97 |
ENSMUST00000074556.7
ENSMUST00000203982.2 |
H2aj
|
H2J.A histone |
| chr7_-_99340830 | 0.95 |
ENSMUST00000208713.2
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr13_+_23868175 | 0.90 |
ENSMUST00000018246.6
|
H2bc4
|
H2B clustered histone 4 |
| chr3_-_96170627 | 0.89 |
ENSMUST00000171473.3
|
H4c14
|
H4 clustered histone 14 |
| chr13_-_21971388 | 0.88 |
ENSMUST00000091751.3
|
H2ac22
|
H2A clustered histone 22 |
| chr17_-_71575584 | 0.87 |
ENSMUST00000233148.2
|
Emilin2
|
elastin microfibril interfacer 2 |
| chr13_-_96572460 | 0.78 |
ENSMUST00000181761.2
|
Ankdd1b
|
ankyrin repeat and death domain containing 1B |
| chr9_-_106533279 | 0.71 |
ENSMUST00000023959.13
ENSMUST00000201681.2 |
Grm2
|
glutamate receptor, metabotropic 2 |
| chr12_+_58258558 | 0.71 |
ENSMUST00000110671.3
ENSMUST00000044299.3 |
Sstr1
|
somatostatin receptor 1 |
| chr7_-_81104423 | 0.69 |
ENSMUST00000178892.3
ENSMUST00000098331.10 |
Cpeb1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr8_+_124532781 | 0.69 |
ENSMUST00000117702.2
|
Rab4a
|
RAB4A, member RAS oncogene family |
| chr13_-_23867924 | 0.67 |
ENSMUST00000171127.4
|
H2ac6
|
H2A clustered histone 6 |
| chr11_+_73131136 | 0.59 |
ENSMUST00000138853.3
|
Trpv1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr4_+_45018583 | 0.56 |
ENSMUST00000133157.8
ENSMUST00000029999.15 ENSMUST00000107814.10 |
Polr1e
|
polymerase (RNA) I polypeptide E |
| chr2_+_177834868 | 0.55 |
ENSMUST00000103065.2
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr11_+_73130872 | 0.54 |
ENSMUST00000108470.8
|
Trpv1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr19_-_47907705 | 0.54 |
ENSMUST00000095998.7
|
Itprip
|
inositol 1,4,5-triphosphate receptor interacting protein |
| chr1_-_128256048 | 0.54 |
ENSMUST00000073490.7
|
Lct
|
lactase |
| chr19_-_47907628 | 0.52 |
ENSMUST00000237029.2
|
Itprip
|
inositol 1,4,5-triphosphate receptor interacting protein |
| chr2_-_119060366 | 0.51 |
ENSMUST00000076084.6
|
Ppp1r14d
|
protein phosphatase 1, regulatory inhibitor subunit 14D |
| chr8_-_85549452 | 0.51 |
ENSMUST00000065539.6
|
Dand5
|
DAN domain family member 5, BMP antagonist |
| chr17_-_43003135 | 0.50 |
ENSMUST00000170723.8
ENSMUST00000164524.2 ENSMUST00000024711.11 ENSMUST00000167993.8 |
Adgrf4
|
adhesion G protein-coupled receptor F4 |
| chr7_+_51530060 | 0.50 |
ENSMUST00000145049.2
|
Gas2
|
growth arrest specific 2 |
| chr7_-_30335277 | 0.49 |
ENSMUST00000108147.3
|
Etv2
|
ets variant 2 |
| chr9_-_123778334 | 0.49 |
ENSMUST00000071404.5
|
Ccr1l1
|
chemokine (C-C motif) receptor 1-like 1 |
| chr9_-_66421868 | 0.48 |
ENSMUST00000056890.10
|
Fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chr6_-_129252323 | 0.48 |
ENSMUST00000204411.2
|
Cd69
|
CD69 antigen |
| chr10_+_33780993 | 0.47 |
ENSMUST00000169670.8
|
Rsph4a
|
radial spoke head 4 homolog A (Chlamydomonas) |
| chr5_+_72360631 | 0.47 |
ENSMUST00000169617.3
|
Atp10d
|
ATPase, class V, type 10D |
| chr1_+_146373352 | 0.45 |
ENSMUST00000132847.8
ENSMUST00000166814.8 |
Brinp3
|
bone morphogenetic protein/retinoic acid inducible neural specific 3 |
| chr9_+_53678801 | 0.44 |
ENSMUST00000048670.10
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr7_-_4550523 | 0.44 |
ENSMUST00000206023.2
|
Syt5
|
synaptotagmin V |
| chrX_+_72454702 | 0.42 |
ENSMUST00000033740.12
|
Zfp92
|
zinc finger protein 92 |
| chr14_+_53775648 | 0.41 |
ENSMUST00000200115.2
ENSMUST00000103650.3 |
Trav12-1
|
T cell receptor alpha variable 12-1 |
| chr11_-_87719115 | 0.39 |
ENSMUST00000136446.3
|
Lpo
|
lactoperoxidase |
| chr4_-_129015493 | 0.39 |
ENSMUST00000135763.2
ENSMUST00000149763.3 ENSMUST00000164649.8 |
Hpca
|
hippocalcin |
| chr7_-_119078330 | 0.38 |
ENSMUST00000207460.2
|
Umod
|
uromodulin |
| chr18_-_77134980 | 0.37 |
ENSMUST00000154665.2
ENSMUST00000026486.13 ENSMUST00000123650.2 ENSMUST00000126153.8 |
Katnal2
|
katanin p60 subunit A-like 2 |
| chr7_-_43885522 | 0.37 |
ENSMUST00000206686.2
ENSMUST00000037220.5 |
1700028J19Rik
|
RIKEN cDNA 1700028J19 gene |
| chr13_-_23015518 | 0.37 |
ENSMUST00000226294.2
ENSMUST00000226180.2 |
Vmn1r210
|
vomeronasal 1 receptor 210 |
| chr13_-_22289994 | 0.37 |
ENSMUST00000227357.2
ENSMUST00000228428.2 |
Vmn1r189
|
vomeronasal 1 receptor 189 |
| chr18_-_77134939 | 0.36 |
ENSMUST00000137354.8
ENSMUST00000137498.8 |
Katnal2
|
katanin p60 subunit A-like 2 |
| chr12_-_76842263 | 0.35 |
ENSMUST00000082431.6
|
Gpx2
|
glutathione peroxidase 2 |
| chr2_-_30084124 | 0.35 |
ENSMUST00000113659.8
ENSMUST00000113660.2 |
Kyat1
|
kynurenine aminotransferase 1 |
| chr1_+_60948149 | 0.34 |
ENSMUST00000027164.9
|
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr13_+_22268610 | 0.33 |
ENSMUST00000228243.2
ENSMUST00000226680.2 |
Vmn1r188
|
vomeronasal 1 receptor 188 |
| chr15_+_81821112 | 0.33 |
ENSMUST00000135663.2
|
Csdc2
|
cold shock domain containing C2, RNA binding |
| chr14_+_64331130 | 0.33 |
ENSMUST00000224112.2
ENSMUST00000165710.2 ENSMUST00000170709.2 |
Prss51
|
protease, serine 51 |
| chr12_+_114594300 | 0.33 |
ENSMUST00000095364.3
|
ENSMUSG00000090765.2
|
|
| chrX_-_143471176 | 0.33 |
ENSMUST00000040184.4
|
Trpc5
|
transient receptor potential cation channel, subfamily C, member 5 |
| chr11_-_82070629 | 0.32 |
ENSMUST00000108189.9
ENSMUST00000021043.5 |
Ccl1
|
chemokine (C-C motif) ligand 1 |
| chr5_+_75735576 | 0.32 |
ENSMUST00000144270.8
ENSMUST00000005815.7 |
Kit
|
KIT proto-oncogene receptor tyrosine kinase |
| chr9_+_108368032 | 0.32 |
ENSMUST00000166103.9
ENSMUST00000085044.14 ENSMUST00000193678.6 ENSMUST00000178075.8 |
Usp19
|
ubiquitin specific peptidase 19 |
| chr18_+_57666852 | 0.32 |
ENSMUST00000079738.10
ENSMUST00000135806.8 ENSMUST00000127130.9 |
Ccdc192
|
coiled-coil domain containing 192 |
| chr9_+_43983493 | 0.32 |
ENSMUST00000176671.2
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr11_+_87457544 | 0.31 |
ENSMUST00000060360.7
|
Septin4
|
septin 4 |
| chr15_-_76906832 | 0.31 |
ENSMUST00000019037.10
ENSMUST00000169226.9 |
Mb
|
myoglobin |
| chr12_-_81468705 | 0.31 |
ENSMUST00000085319.4
|
Adam4
|
a disintegrin and metallopeptidase domain 4 |
| chr5_-_53864874 | 0.30 |
ENSMUST00000031093.5
|
Cckar
|
cholecystokinin A receptor |
| chr9_+_108367801 | 0.30 |
ENSMUST00000006854.13
|
Usp19
|
ubiquitin specific peptidase 19 |
| chr3_-_79645101 | 0.30 |
ENSMUST00000078527.13
|
Rxfp1
|
relaxin/insulin-like family peptide receptor 1 |
| chr7_+_3473178 | 0.30 |
ENSMUST00000108647.3
|
Cacng6
|
calcium channel, voltage-dependent, gamma subunit 6 |
| chr3_+_107538638 | 0.29 |
ENSMUST00000106703.2
|
Gm10961
|
predicted gene 10961 |
| chr7_-_104019046 | 0.29 |
ENSMUST00000106831.3
|
Trim30b
|
tripartite motif-containing 30B |
| chr2_-_173117936 | 0.29 |
ENSMUST00000139306.2
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr1_+_60948307 | 0.29 |
ENSMUST00000097720.4
|
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr14_+_53478202 | 0.28 |
ENSMUST00000179583.3
|
Trav12n-3
|
T cell receptor alpha variable 12N-3 |
| chr14_-_62693735 | 0.28 |
ENSMUST00000165651.8
ENSMUST00000022501.10 |
Gucy1b2
|
guanylate cyclase 1, soluble, beta 2 |
| chr7_-_104018989 | 0.27 |
ENSMUST00000164410.2
|
Trim30b
|
tripartite motif-containing 30B |
| chr12_-_115129697 | 0.27 |
ENSMUST00000103524.2
ENSMUST00000193145.2 |
Ighv8-6
|
immunoglobulin heavy variable V8-6 |
| chr7_+_104506216 | 0.27 |
ENSMUST00000067695.8
|
Usp17la
|
ubiquitin specific peptidase 17-like A |
| chr2_-_17465410 | 0.27 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr1_+_165312738 | 0.26 |
ENSMUST00000111440.8
ENSMUST00000027852.15 ENSMUST00000111439.8 |
Adcy10
|
adenylate cyclase 10 |
| chr3_+_107008343 | 0.26 |
ENSMUST00000197470.5
|
Kcna2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr5_-_72661546 | 0.25 |
ENSMUST00000005352.10
|
Corin
|
corin, serine peptidase |
| chr14_-_50020788 | 0.25 |
ENSMUST00000118129.2
ENSMUST00000036972.14 |
Armh4
|
armadillo-like helical domain containing 4 |
| chr12_+_119909692 | 0.24 |
ENSMUST00000058644.15
|
Tmem196
|
transmembrane protein 196 |
| chr15_-_101471324 | 0.24 |
ENSMUST00000023714.5
|
Krt90
|
keratin 90 |
| chr7_+_103065903 | 0.24 |
ENSMUST00000079348.4
|
Usp17lc
|
ubiquitin specific peptidase 17-like C |
| chr13_+_23398297 | 0.24 |
ENSMUST00000236177.2
|
Vmn1r221
|
vomeronasal 1 receptor 221 |
| chrX_-_94070277 | 0.24 |
ENSMUST00000096367.5
|
Spin4
|
spindlin family, member 4 |
| chr8_+_55053809 | 0.23 |
ENSMUST00000033917.7
|
Spata4
|
spermatogenesis associated 4 |
| chr6_+_120070307 | 0.23 |
ENSMUST00000112711.9
|
Ninj2
|
ninjurin 2 |
| chr10_+_79690452 | 0.23 |
ENSMUST00000165704.8
|
Ptbp1
|
polypyrimidine tract binding protein 1 |
| chr1_+_180720666 | 0.23 |
ENSMUST00000085797.6
|
Lefty2
|
left-right determination factor 2 |
| chr10_+_79690492 | 0.23 |
ENSMUST00000171599.8
ENSMUST00000095457.11 |
Ptbp1
|
polypyrimidine tract binding protein 1 |
| chr12_+_100745314 | 0.22 |
ENSMUST00000069782.11
|
Dglucy
|
D-glutamate cyclase |
| chrX_+_142447286 | 0.22 |
ENSMUST00000112868.8
|
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr6_-_129252396 | 0.22 |
ENSMUST00000032259.6
|
Cd69
|
CD69 antigen |
| chr6_+_90105406 | 0.21 |
ENSMUST00000072859.6
|
Vmn1r51
|
vomeronasal 1 receptor 51 |
| chr11_+_69047815 | 0.21 |
ENSMUST00000036424.3
|
Alox12b
|
arachidonate 12-lipoxygenase, 12R type |
| chr13_+_23343482 | 0.19 |
ENSMUST00000226845.2
ENSMUST00000228666.2 ENSMUST00000227388.2 |
Vmn1r219
|
vomeronasal 1 receptor 219 |
| chr18_-_74340842 | 0.19 |
ENSMUST00000040188.16
|
Ska1
|
spindle and kinetochore associated complex subunit 1 |
| chrX_+_142447361 | 0.18 |
ENSMUST00000126592.8
ENSMUST00000156449.8 ENSMUST00000155215.8 ENSMUST00000112865.8 |
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr11_-_94932158 | 0.18 |
ENSMUST00000038431.8
|
Pdk2
|
pyruvate dehydrogenase kinase, isoenzyme 2 |
| chr16_+_34470290 | 0.17 |
ENSMUST00000148562.8
|
Ropn1
|
ropporin, rhophilin associated protein 1 |
| chr12_+_100745333 | 0.16 |
ENSMUST00000110073.8
ENSMUST00000110069.8 |
Dglucy
|
D-glutamate cyclase |
| chr12_-_114370581 | 0.16 |
ENSMUST00000191918.2
|
Ighv6-4
|
immunoglobulin heavy variable V6-4 |
| chr7_-_12770556 | 0.15 |
ENSMUST00000123541.2
|
Ube2m
|
ubiquitin-conjugating enzyme E2M |
| chr12_-_115031622 | 0.15 |
ENSMUST00000194257.2
|
Ighv8-5
|
immunoglobulin heavy variable V8-5 |
| chr5_+_114912738 | 0.15 |
ENSMUST00000102578.11
|
Ankrd13a
|
ankyrin repeat domain 13a |
| chr13_+_23191826 | 0.15 |
ENSMUST00000228758.2
ENSMUST00000228031.2 ENSMUST00000227573.2 |
Vmn1r213
|
vomeronasal 1 receptor 213 |
| chr19_+_55886708 | 0.15 |
ENSMUST00000148666.3
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr3_+_92281215 | 0.14 |
ENSMUST00000056590.6
|
Sprr2g
|
small proline-rich protein 2G |
| chr1_-_126420659 | 0.14 |
ENSMUST00000161954.8
|
Nckap5
|
NCK-associated protein 5 |
| chr18_+_31767369 | 0.14 |
ENSMUST00000235017.2
ENSMUST00000178164.8 ENSMUST00000025109.8 |
Sap130
|
Sin3A associated protein |
| chr1_+_131616504 | 0.13 |
ENSMUST00000129905.2
|
Rab7b
|
RAB7B, member RAS oncogene family |
| chr14_+_31750946 | 0.13 |
ENSMUST00000022460.11
|
Galnt15
|
polypeptide N-acetylgalactosaminyltransferase 15 |
| chr16_-_44978546 | 0.12 |
ENSMUST00000114600.2
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr1_+_171197660 | 0.12 |
ENSMUST00000111286.9
|
Nectin4
|
nectin cell adhesion molecule 4 |
| chr7_-_127187767 | 0.12 |
ENSMUST00000072155.5
|
Ccdc189
|
coiled-coil domain containing 189 |
| chr7_-_119078472 | 0.12 |
ENSMUST00000209095.2
ENSMUST00000033263.6 ENSMUST00000207261.2 |
Umod
|
uromodulin |
| chr11_+_49414548 | 0.11 |
ENSMUST00000077143.7
|
Olfr1383
|
olfactory receptor 1383 |
| chr1_-_126420420 | 0.11 |
ENSMUST00000162877.8
|
Nckap5
|
NCK-associated protein 5 |
| chr12_-_114217671 | 0.10 |
ENSMUST00000193408.2
|
Ighv3-4
|
immunoglobulin heavy variable V3-4 |
| chr5_+_92540444 | 0.10 |
ENSMUST00000126281.8
|
Art3
|
ADP-ribosyltransferase 3 |
| chr10_-_119075910 | 0.09 |
ENSMUST00000020315.13
|
Cand1
|
cullin associated and neddylation disassociated 1 |
| chr7_+_24920840 | 0.09 |
ENSMUST00000055604.6
|
Zfp526
|
zinc finger protein 526 |
| chr18_-_74340885 | 0.08 |
ENSMUST00000177604.2
|
Ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr6_+_41092970 | 0.08 |
ENSMUST00000103268.3
|
Trbv13-1
|
T cell receptor beta, variable 13-1 |
| chr16_+_90017634 | 0.07 |
ENSMUST00000023707.11
|
Sod1
|
superoxide dismutase 1, soluble |
| chr7_-_24016020 | 0.05 |
ENSMUST00000108436.8
ENSMUST00000032673.15 |
Zfp94
|
zinc finger protein 94 |
| chr3_+_107008867 | 0.05 |
ENSMUST00000038695.6
|
Kcna2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr2_-_164876690 | 0.04 |
ENSMUST00000122070.2
ENSMUST00000121377.8 ENSMUST00000153905.2 ENSMUST00000040381.15 |
Ncoa5
|
nuclear receptor coactivator 5 |
| chr1_-_75187441 | 0.03 |
ENSMUST00000185448.2
|
Glb1l
|
galactosidase, beta 1-like |
| chr14_+_20398230 | 0.03 |
ENSMUST00000224930.2
ENSMUST00000224110.2 ENSMUST00000225942.2 ENSMUST00000051915.7 ENSMUST00000090499.13 ENSMUST00000224721.2 ENSMUST00000090503.12 ENSMUST00000225991.2 ENSMUST00000037698.13 |
Fam149b
|
family with sequence similarity 149, member B |
| chr6_+_41092928 | 0.02 |
ENSMUST00000194399.2
|
Trbv13-1
|
T cell receptor beta, variable 13-1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.3 | 3.9 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.3 | 1.1 | GO:0050960 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.3 | 2.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.2 | 0.9 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.2 | 0.6 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.1 | 0.4 | GO:0031283 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.1 | 0.5 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.1 | 3.8 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 0.5 | GO:0072233 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.1 | 0.3 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 0.1 | 0.3 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 | 0.5 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.1 | 0.6 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.1 | 0.3 | GO:0060618 | nipple development(GO:0060618) |
| 0.1 | 0.5 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.1 | 0.4 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.1 | 1.2 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.1 | 0.3 | GO:1904349 | positive regulation of small intestine smooth muscle contraction(GO:1904349) |
| 0.1 | 0.3 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.1 | 0.7 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.7 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 0.3 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 0.3 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) positive regulation of somatostatin secretion(GO:0090274) |
| 0.0 | 0.2 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.2 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.9 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.8 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.7 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 1.1 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.5 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 1.6 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0048625 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) |
| 0.0 | 0.3 | GO:0003351 | epithelial cilium movement(GO:0003351) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.2 | 2.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.4 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 0.7 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.7 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 3.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.2 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.0 | 0.6 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.4 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.6 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0005009 | insulin-activated receptor activity(GO:0005009) |
| 0.2 | 2.5 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 0.5 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.2 | 1.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.1 | 0.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.7 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 0.3 | GO:0047312 | L-phenylalanine:pyruvate aminotransferase activity(GO:0047312) glutamine-phenylpyruvate transaminase activity(GO:0047316) L-glutamine:pyruvate aminotransferase activity(GO:0047945) |
| 0.1 | 2.1 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 3.8 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 0.5 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.6 | GO:0001179 | RNA polymerase I transcription factor binding(GO:0001179) |
| 0.1 | 0.7 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.1 | 0.2 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.1 | 0.2 | GO:0018169 | ribosomal S6-glutamic acid ligase activity(GO:0018169) |
| 0.0 | 0.6 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.5 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.9 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.7 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.3 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.5 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.9 | GO:0009975 | cyclase activity(GO:0009975) |
| 0.0 | 0.5 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 1.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 1.4 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.7 | GO:0004601 | peroxidase activity(GO:0004601) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.7 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 0.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 2.5 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 0.9 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.6 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |