Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
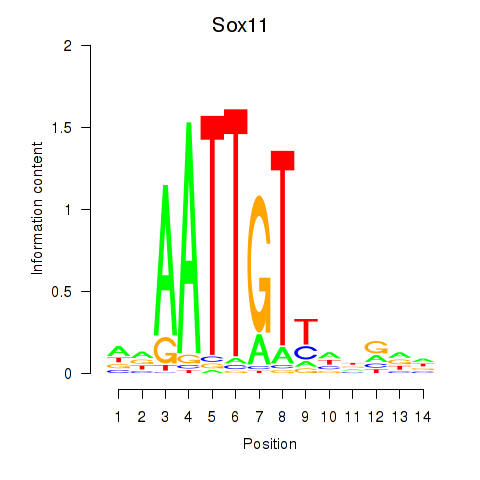
Results for Sox11
Z-value: 0.51
Transcription factors associated with Sox11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox11
|
ENSMUSG00000063632.7 | Sox11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox11 | mm39_v1_chr12_-_27392356_27392573 | 0.57 | 2.8e-04 | Click! |
Activity profile of Sox11 motif
Sorted Z-values of Sox11 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox11
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_60806810 | 4.27 |
ENSMUST00000163779.8
|
Snca
|
synuclein, alpha |
| chr4_-_46404224 | 3.28 |
ENSMUST00000107764.9
|
Hemgn
|
hemogen |
| chr3_-_102074246 | 2.92 |
ENSMUST00000161021.2
|
Vangl1
|
VANGL planar cell polarity 1 |
| chr9_+_65797519 | 2.49 |
ENSMUST00000045802.7
|
Pclaf
|
PCNA clamp associated factor |
| chrX_+_162923474 | 1.54 |
ENSMUST00000073973.11
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr9_-_58648826 | 1.45 |
ENSMUST00000098674.6
|
Rec114
|
REC114 meiotic recombination protein |
| chr5_+_8096074 | 1.28 |
ENSMUST00000088786.11
|
Sri
|
sorcin |
| chrX_+_84617624 | 1.24 |
ENSMUST00000048250.10
ENSMUST00000137438.2 ENSMUST00000146063.2 |
Tab3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr7_-_120673761 | 1.16 |
ENSMUST00000047194.4
|
Igsf6
|
immunoglobulin superfamily, member 6 |
| chr9_+_48073296 | 1.12 |
ENSMUST00000216998.2
ENSMUST00000215780.2 |
Nxpe4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr14_-_20438890 | 1.12 |
ENSMUST00000022345.7
|
Dnajc9
|
DnaJ heat shock protein family (Hsp40) member C9 |
| chr1_-_45542442 | 1.11 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2 |
| chr13_-_58549079 | 1.09 |
ENSMUST00000224524.2
ENSMUST00000224030.2 ENSMUST00000224342.2 |
Hnrnpk
|
heterogeneous nuclear ribonucleoprotein K |
| chr1_+_9918599 | 1.06 |
ENSMUST00000097826.6
|
Sgk3
|
serum/glucocorticoid regulated kinase 3 |
| chr5_+_64969679 | 0.99 |
ENSMUST00000166409.6
ENSMUST00000197879.2 |
Klf3
|
Kruppel-like factor 3 (basic) |
| chrX_+_100317803 | 0.99 |
ENSMUST00000117203.8
ENSMUST00000087948.11 ENSMUST00000087956.6 |
Med12
|
mediator complex subunit 12 |
| chr17_+_43327412 | 0.98 |
ENSMUST00000024708.6
|
Tnfrsf21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr11_+_117006020 | 0.96 |
ENSMUST00000103026.10
ENSMUST00000090433.6 |
Sec14l1
|
SEC14-like lipid binding 1 |
| chr2_+_154390808 | 0.93 |
ENSMUST00000045116.11
ENSMUST00000109709.4 |
1700003F12Rik
|
RIKEN cDNA 1700003F12 gene |
| chr1_+_9918497 | 0.87 |
ENSMUST00000171265.8
|
Sgk3
|
serum/glucocorticoid regulated kinase 3 |
| chr11_-_106084334 | 0.85 |
ENSMUST00000007444.14
ENSMUST00000152008.2 ENSMUST00000103072.10 ENSMUST00000106867.2 |
Strada
|
STE20-related kinase adaptor alpha |
| chr1_-_149798090 | 0.79 |
ENSMUST00000111926.9
|
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr10_-_75946790 | 0.73 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr9_-_111086528 | 0.58 |
ENSMUST00000199404.2
|
Mlh1
|
mutL homolog 1 |
| chr9_+_100956734 | 0.48 |
ENSMUST00000085177.5
|
Msl2
|
MSL complex subunit 2 |
| chr11_+_49327451 | 0.47 |
ENSMUST00000215226.2
|
Olfr1388
|
olfactory receptor 1388 |
| chr3_+_28859585 | 0.47 |
ENSMUST00000043867.11
ENSMUST00000194649.2 |
Rpl22l1
|
ribosomal protein L22 like 1 |
| chr19_-_46958001 | 0.47 |
ENSMUST00000235234.2
|
Nt5c2
|
5'-nucleotidase, cytosolic II |
| chr7_+_108265625 | 0.42 |
ENSMUST00000213979.3
ENSMUST00000216331.2 ENSMUST00000217170.2 |
Olfr510
|
olfactory receptor 510 |
| chr9_-_107512566 | 0.40 |
ENSMUST00000055704.12
|
Gnai2
|
guanine nucleotide binding protein (G protein), alpha inhibiting 2 |
| chr16_-_17729167 | 0.38 |
ENSMUST00000232423.2
ENSMUST00000003621.10 |
Ess2
|
ess-2 splicing factor |
| chr7_-_105386546 | 0.38 |
ENSMUST00000098148.6
|
Rrp8
|
ribosomal RNA processing 8 |
| chr3_+_90201388 | 0.34 |
ENSMUST00000199607.5
|
Gatad2b
|
GATA zinc finger domain containing 2B |
| chr10_+_17672004 | 0.33 |
ENSMUST00000037964.7
|
Txlnb
|
taxilin beta |
| chr2_-_109108618 | 0.31 |
ENSMUST00000081631.10
|
Mettl15
|
methyltransferase like 15 |
| chr5_+_145020910 | 0.31 |
ENSMUST00000124379.3
|
Arpc1a
|
actin related protein 2/3 complex, subunit 1A |
| chr2_+_3115250 | 0.30 |
ENSMUST00000072955.12
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr5_+_145020640 | 0.30 |
ENSMUST00000031625.15
|
Arpc1a
|
actin related protein 2/3 complex, subunit 1A |
| chr9_+_39985125 | 0.29 |
ENSMUST00000054051.5
|
Olfr982
|
olfactory receptor 982 |
| chr4_+_21848039 | 0.29 |
ENSMUST00000098238.9
ENSMUST00000108229.2 |
Pnisr
|
PNN interacting serine/arginine-rich |
| chr19_+_11493825 | 0.28 |
ENSMUST00000163078.8
|
Ms4a6b
|
membrane-spanning 4-domains, subfamily A, member 6B |
| chr10_-_129509659 | 0.28 |
ENSMUST00000213294.2
ENSMUST00000216067.3 ENSMUST00000203424.2 |
Olfr801
|
olfactory receptor 801 |
| chr13_-_113755082 | 0.28 |
ENSMUST00000109241.5
|
Snx18
|
sorting nexin 18 |
| chr4_+_48124902 | 0.26 |
ENSMUST00000107721.8
ENSMUST00000153502.8 ENSMUST00000107720.9 ENSMUST00000064765.6 |
Stx17
|
syntaxin 17 |
| chr7_-_73191484 | 0.25 |
ENSMUST00000197642.2
ENSMUST00000026895.14 ENSMUST00000169922.9 |
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr6_-_69220672 | 0.25 |
ENSMUST00000196201.2
|
Igkv4-71
|
immunoglobulin kappa chain variable 4-71 |
| chr19_-_4675352 | 0.24 |
ENSMUST00000224707.2
ENSMUST00000237059.2 |
Rce1
|
Ras converting CAAX endopeptidase 1 |
| chr18_+_44403169 | 0.24 |
ENSMUST00000042747.4
|
Npy6r
|
neuropeptide Y receptor Y6 |
| chr19_-_4675300 | 0.22 |
ENSMUST00000225264.2
ENSMUST00000237022.2 ENSMUST00000224675.3 |
Rce1
|
Ras converting CAAX endopeptidase 1 |
| chr5_+_144037171 | 0.22 |
ENSMUST00000041804.8
|
Lmtk2
|
lemur tyrosine kinase 2 |
| chr1_-_138103021 | 0.22 |
ENSMUST00000182755.8
ENSMUST00000193650.2 ENSMUST00000182283.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr6_-_87312743 | 0.21 |
ENSMUST00000042025.12
ENSMUST00000205033.2 |
Antxr1
|
anthrax toxin receptor 1 |
| chr18_+_44204457 | 0.21 |
ENSMUST00000068473.4
ENSMUST00000236634.2 |
Spink6
|
serine peptidase inhibitor, Kazal type 6 |
| chr11_-_50183129 | 0.21 |
ENSMUST00000059458.5
|
Maml1
|
mastermind like transcriptional coactivator 1 |
| chr6_+_108771840 | 0.21 |
ENSMUST00000204483.2
|
Arl8b
|
ADP-ribosylation factor-like 8B |
| chr11_-_33942981 | 0.21 |
ENSMUST00000238903.2
|
Kcnip1
|
Kv channel-interacting protein 1 |
| chr15_+_58004793 | 0.20 |
ENSMUST00000227142.2
ENSMUST00000226955.2 |
Wdyhv1
|
WDYHV motif containing 1 |
| chr9_-_19968858 | 0.20 |
ENSMUST00000216538.2
ENSMUST00000212098.3 |
Olfr867
|
olfactory receptor 867 |
| chr9_+_51958453 | 0.19 |
ENSMUST00000163153.9
|
Rdx
|
radixin |
| chr4_-_123033721 | 0.19 |
ENSMUST00000030404.5
|
Ppie
|
peptidylprolyl isomerase E (cyclophilin E) |
| chr8_+_85952617 | 0.19 |
ENSMUST00000218663.2
|
Olfr371
|
olfactory receptor 371 |
| chr4_-_129271909 | 0.18 |
ENSMUST00000030610.3
|
Zbtb8a
|
zinc finger and BTB domain containing 8a |
| chr6_-_69020489 | 0.17 |
ENSMUST00000103342.4
|
Igkv4-79
|
immunoglobulin kappa variable 4-79 |
| chr2_+_111581351 | 0.17 |
ENSMUST00000207590.4
|
Olfr1301
|
olfactory receptor 1301 |
| chr4_+_109092610 | 0.17 |
ENSMUST00000106628.8
|
Calr4
|
calreticulin 4 |
| chr13_-_41640757 | 0.16 |
ENSMUST00000021794.14
|
Nedd9
|
neural precursor cell expressed, developmentally down-regulated gene 9 |
| chr10_+_115405891 | 0.16 |
ENSMUST00000173620.2
|
A930009A15Rik
|
RIKEN cDNA A930009A15 gene |
| chr6_-_69584812 | 0.16 |
ENSMUST00000103359.3
|
Igkv4-55
|
immunoglobulin kappa variable 4-55 |
| chr4_+_109092459 | 0.15 |
ENSMUST00000106631.9
|
Calr4
|
calreticulin 4 |
| chr15_-_80989200 | 0.14 |
ENSMUST00000109579.9
|
Mrtfa
|
myocardin related transcription factor A |
| chr16_+_58967409 | 0.14 |
ENSMUST00000216957.3
|
Olfr195
|
olfactory receptor 195 |
| chr5_-_87638728 | 0.13 |
ENSMUST00000147854.6
|
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr2_-_86109346 | 0.13 |
ENSMUST00000217294.2
ENSMUST00000217245.2 ENSMUST00000216432.2 |
Olfr1051
|
olfactory receptor 1051 |
| chr6_-_42670021 | 0.13 |
ENSMUST00000121083.8
|
Tcaf1
|
TRPM8 channel-associated factor 1 |
| chr7_-_55612213 | 0.13 |
ENSMUST00000032635.14
ENSMUST00000152649.8 |
Nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 homolog (human) |
| chr16_+_33071784 | 0.12 |
ENSMUST00000023502.6
|
Snx4
|
sorting nexin 4 |
| chr2_-_110591909 | 0.12 |
ENSMUST00000140777.2
|
Ano3
|
anoctamin 3 |
| chr1_-_138102972 | 0.12 |
ENSMUST00000195533.6
ENSMUST00000183301.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr8_-_62576140 | 0.11 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr11_+_58549642 | 0.11 |
ENSMUST00000214392.2
|
Olfr322
|
olfactory receptor 322 |
| chr2_+_69050315 | 0.11 |
ENSMUST00000005364.12
ENSMUST00000112317.3 |
G6pc2
|
glucose-6-phosphatase, catalytic, 2 |
| chr10_-_89270554 | 0.10 |
ENSMUST00000220071.2
|
Gas2l3
|
growth arrest-specific 2 like 3 |
| chr5_+_16758897 | 0.10 |
ENSMUST00000196645.2
|
Hgf
|
hepatocyte growth factor |
| chr18_+_37143758 | 0.10 |
ENSMUST00000115657.10
ENSMUST00000192447.6 |
Pcdha11
|
protocadherin alpha 11 |
| chr11_-_116572116 | 0.10 |
ENSMUST00000144398.2
|
St6galnac2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 |
| chr2_-_89195205 | 0.09 |
ENSMUST00000111543.2
ENSMUST00000137692.3 |
Olfr1234
|
olfactory receptor 1234 |
| chr6_+_68279392 | 0.08 |
ENSMUST00000103322.3
|
Igkv2-109
|
immunoglobulin kappa variable 2-109 |
| chr17_+_38456172 | 0.08 |
ENSMUST00000215078.3
ENSMUST00000215549.3 ENSMUST00000173610.2 |
Olfr133
|
olfactory receptor 133 |
| chr6_-_129637519 | 0.07 |
ENSMUST00000119533.2
ENSMUST00000145984.8 ENSMUST00000118401.8 ENSMUST00000112057.9 ENSMUST00000071920.11 |
Klrc2
|
killer cell lectin-like receptor subfamily C, member 2 |
| chr13_-_95615229 | 0.07 |
ENSMUST00000022186.5
|
S100z
|
S100 calcium binding protein, zeta |
| chr4_+_152160713 | 0.07 |
ENSMUST00000239025.2
|
Plekhg5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr2_-_86589298 | 0.07 |
ENSMUST00000215991.2
ENSMUST00000217043.3 |
Olfr1090
|
olfactory receptor 1090 |
| chr18_-_35841435 | 0.07 |
ENSMUST00000236738.2
ENSMUST00000237995.2 |
Dnajc18
|
DnaJ heat shock protein family (Hsp40) member C18 |
| chr12_-_113733922 | 0.07 |
ENSMUST00000180013.3
|
Ighv2-9-1
|
immunoglobulin heavy variable 2-9-1 |
| chr10_-_129741736 | 0.07 |
ENSMUST00000216182.2
|
Olfr815
|
olfactory receptor 815 |
| chr13_+_24925192 | 0.06 |
ENSMUST00000136906.2
|
Armh2
|
armadillo-like helical domain containing 2 |
| chr2_+_121991181 | 0.06 |
ENSMUST00000036089.8
|
Trim69
|
tripartite motif-containing 69 |
| chr1_+_92545510 | 0.05 |
ENSMUST00000213247.2
|
Olfr12
|
olfactory receptor 12 |
| chr8_+_106154266 | 0.05 |
ENSMUST00000067305.8
|
Lrrc36
|
leucine rich repeat containing 36 |
| chr4_+_33132502 | 0.05 |
ENSMUST00000029947.6
|
Gabrr1
|
gamma-aminobutyric acid (GABA) C receptor, subunit rho 1 |
| chr10_+_5589210 | 0.05 |
ENSMUST00000019906.6
|
Vip
|
vasoactive intestinal polypeptide |
| chr12_-_98700886 | 0.05 |
ENSMUST00000085116.4
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr13_+_23431304 | 0.05 |
ENSMUST00000235331.2
ENSMUST00000236495.2 ENSMUST00000238002.2 |
Vmn1r223
|
vomeronasal 1 receptor 223 |
| chr1_-_156168522 | 0.04 |
ENSMUST00000212747.2
|
Axdnd1
|
axonemal dynein light chain domain containing 1 |
| chr2_-_85442212 | 0.04 |
ENSMUST00000216571.2
ENSMUST00000213837.2 |
Olfr1000
|
olfactory receptor 1000 |
| chr2_-_79287095 | 0.04 |
ENSMUST00000041099.5
|
Neurod1
|
neurogenic differentiation 1 |
| chr7_-_85986774 | 0.04 |
ENSMUST00000213705.2
ENSMUST00000217494.2 |
Olfr307
|
olfactory receptor 307 |
| chr18_+_37580692 | 0.03 |
ENSMUST00000052387.5
|
Pcdhb14
|
protocadherin beta 14 |
| chr6_+_116531196 | 0.03 |
ENSMUST00000217313.2
ENSMUST00000214699.2 ENSMUST00000215846.2 ENSMUST00000203867.2 |
Olfr214
|
olfactory receptor 214 |
| chr6_-_68968278 | 0.03 |
ENSMUST00000197966.2
|
Igkv4-81
|
immunoglobulin kappa variable 4-81 |
| chr11_+_74077075 | 0.03 |
ENSMUST00000206114.3
|
Olfr403
|
olfactory receptor 403 |
| chr1_-_173948728 | 0.03 |
ENSMUST00000214446.2
|
Olfr231
|
olfactory receptor 231 |
| chr13_-_22689551 | 0.02 |
ENSMUST00000228020.2
|
Vmn1r202
|
vomeronasal 1 receptor 202 |
| chr6_+_56926317 | 0.02 |
ENSMUST00000227073.2
|
Vmn1r4
|
vomeronasal 1 receptor 4 |
| chr11_-_41891662 | 0.02 |
ENSMUST00000070725.11
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
| chr17_+_35827997 | 0.02 |
ENSMUST00000164242.9
ENSMUST00000045956.14 |
Cchcr1
|
coiled-coil alpha-helical rod protein 1 |
| chr10_-_8638215 | 0.02 |
ENSMUST00000212553.2
|
Sash1
|
SAM and SH3 domain containing 1 |
| chr5_-_86893645 | 0.01 |
ENSMUST00000161306.2
|
Tmprss11e
|
transmembrane protease, serine 11e |
| chr4_+_135455427 | 0.01 |
ENSMUST00000102546.4
|
Il22ra1
|
interleukin 22 receptor, alpha 1 |
| chr3_-_75359125 | 0.00 |
ENSMUST00000204341.3
|
Wdr49
|
WD repeat domain 49 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.4 | 1.5 | GO:0015827 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.3 | 1.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 1.0 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.2 | 1.1 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.2 | 2.5 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 0.6 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) meiotic metaphase plate congression(GO:0051311) |
| 0.1 | 1.3 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.1 | 1.0 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 1.0 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 0.8 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.5 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.1 | 0.3 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.1 | 0.3 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.1 | 0.2 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.1 | 0.7 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.4 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.3 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.3 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 1.6 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.4 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.2 | GO:1902966 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 | 0.1 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 2.9 | GO:0043473 | pigmentation(GO:0043473) |
| 0.0 | 0.2 | GO:1900004 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 3.3 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.0 | 0.2 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.1 | GO:0061591 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 1.1 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.5 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.1 | GO:0002441 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.3 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.4 | 1.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.3 | 1.3 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.2 | 0.6 | GO:0005715 | chiasma(GO:0005712) late recombination nodule(GO:0005715) |
| 0.1 | 0.6 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.5 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.4 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.3 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 2.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 1.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.8 | GO:0042588 | zymogen granule(GO:0042588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.3 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.3 | 1.5 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.6 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.1 | 1.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 0.8 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 1.8 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 0.3 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.7 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 1.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0001601 | peptide YY receptor activity(GO:0001601) pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 1.0 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 1.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.0 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.9 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.8 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.9 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.9 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.8 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 1.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 2.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |