Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
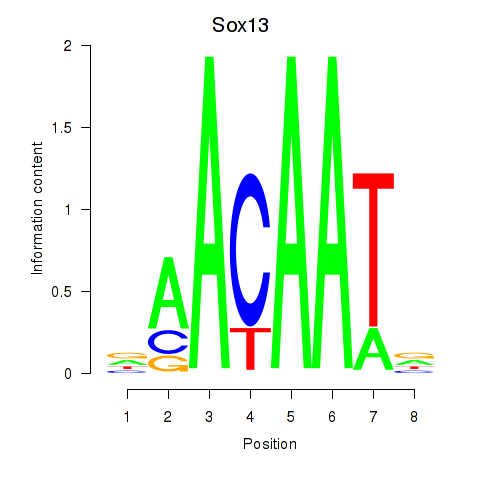
Results for Sox13
Z-value: 0.32
Transcription factors associated with Sox13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox13
|
ENSMUSG00000070643.12 | Sox13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox13 | mm39_v1_chr1_-_133352115_133352138 | 0.53 | 7.9e-04 | Click! |
Activity profile of Sox13 motif
Sorted Z-values of Sox13 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox13
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_99328969 | 1.87 |
ENSMUST00000017743.3
|
Krt20
|
keratin 20 |
| chr19_+_44282113 | 1.45 |
ENSMUST00000026221.7
|
Scd2
|
stearoyl-Coenzyme A desaturase 2 |
| chr7_+_121818692 | 0.65 |
ENSMUST00000033152.5
|
Chp2
|
calcineurin-like EF hand protein 2 |
| chr1_+_172327812 | 0.53 |
ENSMUST00000192460.2
|
Tagln2
|
transgelin 2 |
| chr5_+_146769700 | 0.44 |
ENSMUST00000035983.12
|
Rpl21
|
ribosomal protein L21 |
| chr6_-_87312743 | 0.44 |
ENSMUST00000042025.12
ENSMUST00000205033.2 |
Antxr1
|
anthrax toxin receptor 1 |
| chr18_-_6516089 | 0.39 |
ENSMUST00000115870.9
|
Epc1
|
enhancer of polycomb homolog 1 |
| chr8_+_108271643 | 0.39 |
ENSMUST00000212543.2
|
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr4_+_103000248 | 0.30 |
ENSMUST00000106855.2
|
Mier1
|
MEIR1 treanscription regulator |
| chr9_-_57673128 | 0.25 |
ENSMUST00000065330.8
|
Clk3
|
CDC-like kinase 3 |
| chr18_-_43610829 | 0.25 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr3_-_75177378 | 0.24 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr11_+_29668563 | 0.23 |
ENSMUST00000060992.6
|
Rtn4
|
reticulon 4 |
| chr15_+_6416079 | 0.22 |
ENSMUST00000080880.12
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr2_+_4886298 | 0.08 |
ENSMUST00000027973.14
|
Sephs1
|
selenophosphate synthetase 1 |
| chr19_+_41471067 | 0.06 |
ENSMUST00000067795.13
|
Lcor
|
ligand dependent nuclear receptor corepressor |
| chr7_-_84328553 | 0.06 |
ENSMUST00000069537.3
ENSMUST00000207865.2 ENSMUST00000178385.9 ENSMUST00000208782.2 |
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr13_-_63712349 | 0.05 |
ENSMUST00000192155.6
|
Ptch1
|
patched 1 |
| chr1_+_39940043 | 0.03 |
ENSMUST00000168431.7
ENSMUST00000163854.9 |
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr6_-_87312681 | 0.03 |
ENSMUST00000204805.3
|
Antxr1
|
anthrax toxin receptor 1 |
| chr14_-_121935829 | 0.03 |
ENSMUST00000040700.9
ENSMUST00000212181.2 |
Dock9
|
dedicator of cytokinesis 9 |
| chrX_+_132751729 | 0.02 |
ENSMUST00000033602.9
|
Tnmd
|
tenomodulin |
| chr8_-_62576140 | 0.01 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr6_-_122317457 | 0.00 |
ENSMUST00000160843.8
|
Phc1
|
polyhomeotic 1 |
| chr19_+_41471395 | 0.00 |
ENSMUST00000237208.2
ENSMUST00000238398.2 |
Lcor
|
ligand dependent nuclear receptor corepressor |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.5 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 | 1.9 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.2 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.6 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.3 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 1.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.1 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |