Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Sox18_Sox12
Z-value: 2.11
Transcription factors associated with Sox18_Sox12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
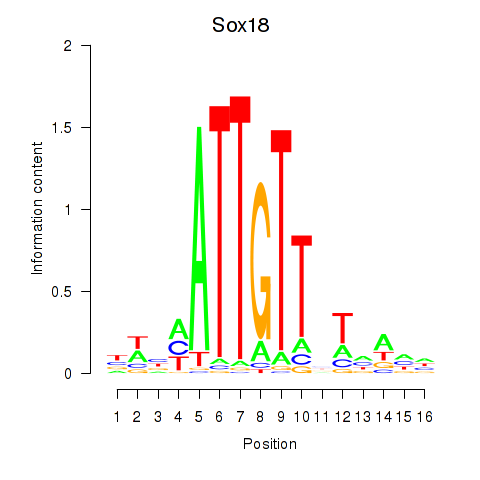
Sox18
|
ENSMUSG00000046470.6 | Sox18 |
|
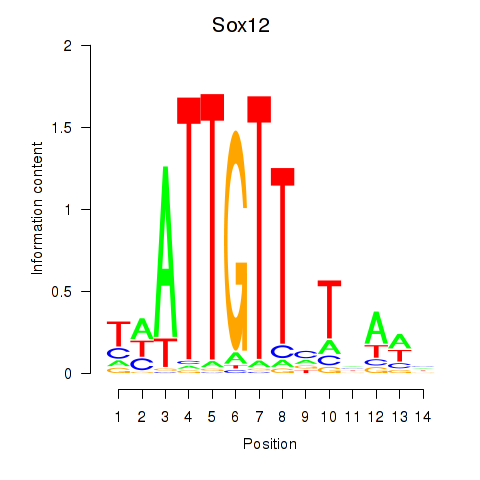
Sox12
|
ENSMUSG00000051817.9 | Sox12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox12 | mm39_v1_chr2_-_152239966_152239997 | -0.90 | 1.8e-13 | Click! |
| Sox18 | mm39_v1_chr2_-_181313415_181313438 | -0.58 | 1.8e-04 | Click! |
Activity profile of Sox18_Sox12 motif
Sorted Z-values of Sox18_Sox12 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox18_Sox12
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_40078132 | 12.96 |
ENSMUST00000068094.13
ENSMUST00000080171.3 |
Cyp2c50
|
cytochrome P450, family 2, subfamily c, polypeptide 50 |
| chr2_+_102489558 | 9.98 |
ENSMUST00000111213.8
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr2_+_102536701 | 8.53 |
ENSMUST00000123759.8
ENSMUST00000005220.11 ENSMUST00000111212.8 |
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr19_-_40062174 | 7.60 |
ENSMUST00000048959.5
|
Cyp2c54
|
cytochrome P450, family 2, subfamily c, polypeptide 54 |
| chr4_+_134123631 | 7.36 |
ENSMUST00000105869.9
|
Pafah2
|
platelet-activating factor acetylhydrolase 2 |
| chr3_-_113371392 | 7.23 |
ENSMUST00000067980.12
|
Amy1
|
amylase 1, salivary |
| chr5_-_87074380 | 6.91 |
ENSMUST00000031183.3
|
Ugt2b1
|
UDP glucuronosyltransferase 2 family, polypeptide B1 |
| chr1_-_72251466 | 6.90 |
ENSMUST00000048860.9
|
Mreg
|
melanoregulin |
| chr13_+_4099001 | 6.82 |
ENSMUST00000118717.10
|
Akr1c14
|
aldo-keto reductase family 1, member C14 |
| chr19_-_8382424 | 6.22 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr18_-_75094323 | 6.06 |
ENSMUST00000066532.5
|
Lipg
|
lipase, endothelial |
| chr1_-_130589349 | 5.81 |
ENSMUST00000027657.14
|
C4bp
|
complement component 4 binding protein |
| chr7_+_127399776 | 5.47 |
ENSMUST00000046863.12
ENSMUST00000206674.2 ENSMUST00000106272.8 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr1_-_130589321 | 5.13 |
ENSMUST00000137276.3
|
C4bp
|
complement component 4 binding protein |
| chr19_-_7779943 | 4.97 |
ENSMUST00000120522.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr9_+_74883377 | 4.95 |
ENSMUST00000081746.7
|
Fam214a
|
family with sequence similarity 214, member A |
| chr19_-_7780025 | 4.55 |
ENSMUST00000065634.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr16_+_22739028 | 4.20 |
ENSMUST00000232097.2
|
Fetub
|
fetuin beta |
| chr19_-_8109346 | 4.10 |
ENSMUST00000065651.5
|
Slc22a28
|
solute carrier family 22, member 28 |
| chr10_-_127843377 | 3.81 |
ENSMUST00000219447.2
ENSMUST00000219780.2 ENSMUST00000219707.2 ENSMUST00000219953.2 ENSMUST00000219183.2 |
Hsd17b6
|
hydroxysteroid (17-beta) dehydrogenase 6 |
| chr1_-_140111138 | 3.79 |
ENSMUST00000111976.9
ENSMUST00000066859.13 |
Cfh
|
complement component factor h |
| chr13_-_55574582 | 3.77 |
ENSMUST00000170921.2
|
F12
|
coagulation factor XII (Hageman factor) |
| chr6_+_42222841 | 3.72 |
ENSMUST00000031897.8
|
Gstk1
|
glutathione S-transferase kappa 1 |
| chr1_+_67162176 | 3.72 |
ENSMUST00000027144.8
|
Cps1
|
carbamoyl-phosphate synthetase 1 |
| chr13_-_55574596 | 3.61 |
ENSMUST00000021948.15
|
F12
|
coagulation factor XII (Hageman factor) |
| chr2_+_121978156 | 3.60 |
ENSMUST00000102476.5
|
B2m
|
beta-2 microglobulin |
| chr14_-_52151537 | 3.59 |
ENSMUST00000227402.2
ENSMUST00000227237.2 |
Ndrg2
|
N-myc downstream regulated gene 2 |
| chr1_-_140111018 | 3.52 |
ENSMUST00000192880.6
ENSMUST00000111977.8 |
Cfh
|
complement component factor h |
| chr6_-_141892686 | 3.49 |
ENSMUST00000042119.6
|
Slco1a1
|
solute carrier organic anion transporter family, member 1a1 |
| chr12_+_40495951 | 3.43 |
ENSMUST00000037488.8
|
Dock4
|
dedicator of cytokinesis 4 |
| chr19_+_4761181 | 3.39 |
ENSMUST00000008991.8
|
Sptbn2
|
spectrin beta, non-erythrocytic 2 |
| chr6_-_141892517 | 3.28 |
ENSMUST00000168119.8
|
Slco1a1
|
solute carrier organic anion transporter family, member 1a1 |
| chr9_-_36708569 | 3.26 |
ENSMUST00000163192.11
|
Ei24
|
etoposide induced 2.4 mRNA |
| chr9_-_36708599 | 3.16 |
ENSMUST00000238932.2
ENSMUST00000115086.13 |
Ei24
|
etoposide induced 2.4 mRNA |
| chr7_-_25239229 | 3.11 |
ENSMUST00000044547.10
ENSMUST00000066503.14 ENSMUST00000064862.13 |
Ceacam2
|
carcinoembryonic antigen-related cell adhesion molecule 2 |
| chr3_+_63203235 | 3.08 |
ENSMUST00000194134.6
|
Mme
|
membrane metallo endopeptidase |
| chr11_+_101258368 | 3.07 |
ENSMUST00000019469.3
|
G6pc
|
glucose-6-phosphatase, catalytic |
| chr1_+_176642226 | 3.07 |
ENSMUST00000056773.15
ENSMUST00000027785.15 |
Sdccag8
|
serologically defined colon cancer antigen 8 |
| chr8_-_93956143 | 3.01 |
ENSMUST00000176282.2
ENSMUST00000034173.14 |
Ces1e
|
carboxylesterase 1E |
| chr17_-_59320257 | 2.85 |
ENSMUST00000174122.2
ENSMUST00000025065.12 |
Nudt12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
| chr3_-_113367891 | 2.81 |
ENSMUST00000142505.9
|
Amy1
|
amylase 1, salivary |
| chr16_-_91415873 | 2.80 |
ENSMUST00000143058.2
ENSMUST00000049244.10 ENSMUST00000169982.2 ENSMUST00000133731.2 |
Dnajc28
|
DnaJ heat shock protein family (Hsp40) member C28 |
| chr2_+_172994841 | 2.78 |
ENSMUST00000029017.6
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chrM_+_10167 | 2.74 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr8_+_3543131 | 2.72 |
ENSMUST00000061508.8
ENSMUST00000207318.2 |
Zfp358
|
zinc finger protein 358 |
| chr14_+_63075127 | 2.63 |
ENSMUST00000014691.10
|
Wdfy2
|
WD repeat and FYVE domain containing 2 |
| chr1_-_133849131 | 2.56 |
ENSMUST00000048432.6
|
Prelp
|
proline arginine-rich end leucine-rich repeat |
| chr6_+_122285615 | 2.51 |
ENSMUST00000007602.15
ENSMUST00000112610.2 |
M6pr
|
mannose-6-phosphate receptor, cation dependent |
| chr3_+_79792238 | 2.44 |
ENSMUST00000135021.2
|
Gask1b
|
golgi associated kinase 1B |
| chr8_-_110305672 | 2.43 |
ENSMUST00000074898.8
|
Hp
|
haptoglobin |
| chrM_+_9870 | 2.39 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr3_+_67337429 | 2.30 |
ENSMUST00000077271.9
|
Gfm1
|
G elongation factor, mitochondrial 1 |
| chrX_+_99811325 | 2.22 |
ENSMUST00000000901.13
ENSMUST00000113736.9 ENSMUST00000087984.11 |
Dlg3
|
discs large MAGUK scaffold protein 3 |
| chr18_-_56705960 | 2.22 |
ENSMUST00000174518.8
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr1_+_44159106 | 2.21 |
ENSMUST00000114709.3
ENSMUST00000129068.2 |
Bivm
|
basic, immunoglobulin-like variable motif containing |
| chrX_+_20416019 | 2.16 |
ENSMUST00000023832.7
|
Rgn
|
regucalcin |
| chr9_-_29323032 | 2.16 |
ENSMUST00000115236.2
|
Ntm
|
neurotrimin |
| chr6_-_47790272 | 2.13 |
ENSMUST00000077290.9
|
Pdia4
|
protein disulfide isomerase associated 4 |
| chr13_-_25204272 | 2.13 |
ENSMUST00000021772.4
|
Mrs2
|
MRS2 magnesium transporter |
| chr5_-_87739442 | 2.12 |
ENSMUST00000031201.9
|
Sult1e1
|
sulfotransferase family 1E, member 1 |
| chr4_-_129132963 | 2.11 |
ENSMUST00000097873.10
|
C77080
|
expressed sequence C77080 |
| chr3_+_79793237 | 2.09 |
ENSMUST00000029567.9
|
Gask1b
|
golgi associated kinase 1B |
| chr7_-_103320398 | 2.08 |
ENSMUST00000062144.4
|
Olfr624
|
olfactory receptor 624 |
| chrX_+_139808351 | 2.03 |
ENSMUST00000033806.5
|
Vsig1
|
V-set and immunoglobulin domain containing 1 |
| chr8_-_45747883 | 2.03 |
ENSMUST00000026907.6
|
Klkb1
|
kallikrein B, plasma 1 |
| chr6_+_121709891 | 1.82 |
ENSMUST00000204124.2
|
Gm7298
|
predicted gene 7298 |
| chr2_+_126394327 | 1.80 |
ENSMUST00000061491.14
|
Slc27a2
|
solute carrier family 27 (fatty acid transporter), member 2 |
| chrX_+_138464065 | 1.80 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr15_+_39609320 | 1.78 |
ENSMUST00000227368.2
ENSMUST00000228556.2 ENSMUST00000022913.6 ENSMUST00000228701.2 ENSMUST00000227792.2 |
Dcstamp
|
dendrocyte expressed seven transmembrane protein |
| chrX_+_141011173 | 1.76 |
ENSMUST00000112914.8
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr14_-_34032311 | 1.74 |
ENSMUST00000111917.3
ENSMUST00000228704.2 |
Shld2
|
shieldin complex subunit 2 |
| chr15_+_31602252 | 1.72 |
ENSMUST00000042702.7
ENSMUST00000161061.3 |
Atpsckmt
|
ATP synthase C subunit lysine N-methyltransferase |
| chr1_+_165616315 | 1.69 |
ENSMUST00000161559.3
|
Cd247
|
CD247 antigen |
| chr2_-_160701523 | 1.67 |
ENSMUST00000103112.8
|
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr13_+_24511387 | 1.67 |
ENSMUST00000224953.2
ENSMUST00000050859.13 ENSMUST00000167746.8 ENSMUST00000224819.2 |
Cmah
|
cytidine monophospho-N-acetylneuraminic acid hydroxylase |
| chrM_+_11735 | 1.66 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr11_+_4823951 | 1.65 |
ENSMUST00000038570.9
|
Nipsnap1
|
nipsnap homolog 1 |
| chr1_-_169575203 | 1.64 |
ENSMUST00000027991.12
ENSMUST00000111357.2 |
Rgs4
|
regulator of G-protein signaling 4 |
| chr3_+_89366425 | 1.63 |
ENSMUST00000029564.12
|
Pmvk
|
phosphomevalonate kinase |
| chr9_-_121745354 | 1.58 |
ENSMUST00000062474.5
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr1_+_171041583 | 1.58 |
ENSMUST00000111328.8
|
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr13_+_93810911 | 1.55 |
ENSMUST00000048001.8
|
Dmgdh
|
dimethylglycine dehydrogenase precursor |
| chr17_+_53873964 | 1.55 |
ENSMUST00000000724.15
|
Kat2b
|
K(lysine) acetyltransferase 2B |
| chr7_-_140462187 | 1.52 |
ENSMUST00000211179.2
|
Sirt3
|
sirtuin 3 |
| chr1_+_171041539 | 1.51 |
ENSMUST00000005820.11
ENSMUST00000075469.12 ENSMUST00000155126.8 |
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr7_-_140462221 | 1.51 |
ENSMUST00000026559.14
|
Sirt3
|
sirtuin 3 |
| chr13_-_63036096 | 1.47 |
ENSMUST00000092888.11
|
Fbp1
|
fructose bisphosphatase 1 |
| chr19_-_34504871 | 1.43 |
ENSMUST00000178114.2
ENSMUST00000049572.15 |
Lipa
|
lysosomal acid lipase A |
| chrX_+_141010919 | 1.43 |
ENSMUST00000042329.12
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr13_-_14237958 | 1.41 |
ENSMUST00000223174.2
|
Ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr5_-_31265562 | 1.41 |
ENSMUST00000201396.2
ENSMUST00000202740.4 |
Slc30a3
|
solute carrier family 30 (zinc transporter), member 3 |
| chrM_+_14138 | 1.37 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr1_+_52884172 | 1.35 |
ENSMUST00000159352.8
ENSMUST00000044478.7 |
Hibch
|
3-hydroxyisobutyryl-Coenzyme A hydrolase |
| chr15_-_100576715 | 1.34 |
ENSMUST00000229869.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr3_+_95801263 | 1.32 |
ENSMUST00000015894.12
|
Aph1a
|
aph1 homolog A, gamma secretase subunit |
| chrM_+_9459 | 1.32 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr4_-_119217079 | 1.30 |
ENSMUST00000143494.3
ENSMUST00000154606.9 |
Ccdc30
|
coiled-coil domain containing 30 |
| chr4_-_49549489 | 1.28 |
ENSMUST00000029987.10
|
Aldob
|
aldolase B, fructose-bisphosphate |
| chr13_+_14238361 | 1.28 |
ENSMUST00000129488.8
ENSMUST00000110536.8 ENSMUST00000110534.8 ENSMUST00000039538.15 ENSMUST00000110533.2 |
Arid4b
|
AT rich interactive domain 4B (RBP1-like) |
| chr7_+_99659121 | 1.28 |
ENSMUST00000107084.8
|
Chrdl2
|
chordin-like 2 |
| chr13_+_113171645 | 1.24 |
ENSMUST00000180543.8
ENSMUST00000181568.8 ENSMUST00000109244.9 ENSMUST00000181117.8 ENSMUST00000181741.2 |
Cdc20b
|
cell division cycle 20B |
| chr1_-_46927230 | 1.22 |
ENSMUST00000185520.2
|
Slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr17_+_37083802 | 1.22 |
ENSMUST00000041531.7
|
H2-M10.5
|
histocompatibility 2, M region locus 10.5 |
| chr17_-_40553176 | 1.21 |
ENSMUST00000026499.6
|
Crisp3
|
cysteine-rich secretory protein 3 |
| chr13_-_93810808 | 1.21 |
ENSMUST00000015941.8
|
Bhmt2
|
betaine-homocysteine methyltransferase 2 |
| chr14_-_34032450 | 1.16 |
ENSMUST00000227375.2
|
Shld2
|
shieldin complex subunit 2 |
| chr10_+_80084955 | 1.15 |
ENSMUST00000105364.8
|
Ndufs7
|
NADH:ubiquinone oxidoreductase core subunit S7 |
| chr19_-_8196196 | 1.15 |
ENSMUST00000113298.9
|
Slc22a29
|
solute carrier family 22. member 29 |
| chr9_-_39918243 | 1.13 |
ENSMUST00000073932.4
|
Olfr980
|
olfactory receptor 980 |
| chr5_+_14564932 | 1.10 |
ENSMUST00000182407.8
ENSMUST00000030691.17 |
Pclo
|
piccolo (presynaptic cytomatrix protein) |
| chrM_-_14061 | 1.09 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr4_-_148234123 | 1.08 |
ENSMUST00000126615.8
|
Fbxo6
|
F-box protein 6 |
| chr12_-_76224025 | 1.07 |
ENSMUST00000101291.11
ENSMUST00000218621.2 ENSMUST00000076634.5 |
Esr2
|
estrogen receptor 2 (beta) |
| chr10_-_12689345 | 1.07 |
ENSMUST00000217899.2
|
Utrn
|
utrophin |
| chr12_+_44268134 | 1.06 |
ENSMUST00000122902.8
|
Pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr18_+_21134302 | 1.06 |
ENSMUST00000234107.2
ENSMUST00000072847.12 |
Rnf138
|
ring finger protein 138 |
| chr1_+_131794962 | 1.05 |
ENSMUST00000112386.8
ENSMUST00000027693.8 |
Rab29
|
RAB29, member RAS oncogene family |
| chr12_-_40495753 | 1.02 |
ENSMUST00000069692.10
ENSMUST00000069637.15 |
Zfp277
|
zinc finger protein 277 |
| chr3_+_95801325 | 1.01 |
ENSMUST00000197081.2
ENSMUST00000056710.10 |
Aph1a
|
aph1 homolog A, gamma secretase subunit |
| chrX_-_84820250 | 1.01 |
ENSMUST00000113978.9
|
Gk
|
glycerol kinase |
| chr5_-_120610828 | 1.01 |
ENSMUST00000052258.14
ENSMUST00000031594.13 |
Sdsl
|
serine dehydratase-like |
| chr8_-_71315902 | 0.99 |
ENSMUST00000212611.2
|
Kcnn1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
| chr4_+_60003438 | 0.99 |
ENSMUST00000107517.8
ENSMUST00000107520.2 |
Mup6
|
major urinary protein 6 |
| chr14_+_11307729 | 0.98 |
ENSMUST00000160956.2
ENSMUST00000160340.8 ENSMUST00000162278.8 |
Fhit
|
fragile histidine triad gene |
| chr7_+_18962252 | 0.97 |
ENSMUST00000063976.9
|
Opa3
|
optic atrophy 3 |
| chr9_+_107765320 | 0.96 |
ENSMUST00000191906.6
ENSMUST00000035202.4 |
Mon1a
|
MON1 homolog A, secretory traffciking associated |
| chr9_-_29323500 | 0.95 |
ENSMUST00000115237.8
|
Ntm
|
neurotrimin |
| chr3_+_63202940 | 0.95 |
ENSMUST00000194150.6
|
Mme
|
membrane metallo endopeptidase |
| chr3_+_59939175 | 0.93 |
ENSMUST00000029325.5
|
Aadac
|
arylacetamide deacetylase |
| chr5_-_91550853 | 0.93 |
ENSMUST00000121044.6
|
Btc
|
betacellulin, epidermal growth factor family member |
| chr3_+_108561247 | 0.92 |
ENSMUST00000124384.8
ENSMUST00000029483.15 |
Clcc1
|
chloride channel CLIC-like 1 |
| chr3_-_57599956 | 0.90 |
ENSMUST00000238789.2
ENSMUST00000197088.5 ENSMUST00000099091.4 |
Ankub1
|
ankrin repeat and ubiquitin domain containing 1 |
| chrM_+_7779 | 0.89 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr1_+_165616250 | 0.89 |
ENSMUST00000161971.8
ENSMUST00000187313.7 ENSMUST00000178336.8 ENSMUST00000005907.12 ENSMUST00000027849.11 |
Cd247
|
CD247 antigen |
| chr9_-_48391838 | 0.88 |
ENSMUST00000216470.2
ENSMUST00000217037.2 ENSMUST00000034524.5 ENSMUST00000213895.2 |
Rexo2
|
RNA exonuclease 2 |
| chr18_+_69477541 | 0.88 |
ENSMUST00000114985.10
ENSMUST00000128706.8 ENSMUST00000201781.4 ENSMUST00000202674.4 |
Tcf4
|
transcription factor 4 |
| chr13_+_63387870 | 0.88 |
ENSMUST00000159152.3
ENSMUST00000221820.2 |
Aopep
|
aminopeptidase O |
| chr16_+_56024676 | 0.88 |
ENSMUST00000160116.8
ENSMUST00000069936.8 |
Impg2
|
interphotoreceptor matrix proteoglycan 2 |
| chr9_+_7692087 | 0.87 |
ENSMUST00000018767.8
|
Mmp7
|
matrix metallopeptidase 7 |
| chr8_-_68363564 | 0.86 |
ENSMUST00000093468.12
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr17_+_12803019 | 0.86 |
ENSMUST00000046959.9
ENSMUST00000233066.2 |
Slc22a2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chr16_-_23339329 | 0.85 |
ENSMUST00000230040.2
ENSMUST00000229619.2 |
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr10_+_34359395 | 0.84 |
ENSMUST00000019913.15
|
Frk
|
fyn-related kinase |
| chr2_-_87322762 | 0.84 |
ENSMUST00000026957.4
|
Pramel7
|
PRAME like 7 |
| chr3_+_96543143 | 0.82 |
ENSMUST00000165842.3
|
Pex11b
|
peroxisomal biogenesis factor 11 beta |
| chr10_-_121146940 | 0.81 |
ENSMUST00000064107.7
|
Tbc1d30
|
TBC1 domain family, member 30 |
| chr13_+_47276132 | 0.81 |
ENSMUST00000068891.12
|
Rnf144b
|
ring finger protein 144B |
| chr15_-_65784246 | 0.81 |
ENSMUST00000060522.11
|
Oc90
|
otoconin 90 |
| chr17_-_18498018 | 0.80 |
ENSMUST00000172190.4
ENSMUST00000231815.3 |
Vmn2r94
|
vomeronasal 2, receptor 94 |
| chr13_+_63387827 | 0.80 |
ENSMUST00000222929.2
|
Aopep
|
aminopeptidase O |
| chr2_+_158508609 | 0.80 |
ENSMUST00000103116.10
|
Ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr11_-_42073737 | 0.79 |
ENSMUST00000206085.2
ENSMUST00000020707.12 ENSMUST00000132971.3 |
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr2_+_15060051 | 0.77 |
ENSMUST00000069870.11
ENSMUST00000239125.2 ENSMUST00000193836.3 |
Arl5b
|
ADP-ribosylation factor-like 5B |
| chr15_-_65784103 | 0.77 |
ENSMUST00000079776.14
|
Oc90
|
otoconin 90 |
| chr14_-_68893253 | 0.77 |
ENSMUST00000225767.3
ENSMUST00000111072.8 ENSMUST00000022642.6 ENSMUST00000224039.2 |
Adam28
|
a disintegrin and metallopeptidase domain 28 |
| chr6_+_54241830 | 0.77 |
ENSMUST00000146114.8
|
Chn2
|
chimerin 2 |
| chr2_+_36575800 | 0.77 |
ENSMUST00000213258.2
|
Olfr346
|
olfactory receptor 346 |
| chr11_+_108811626 | 0.75 |
ENSMUST00000140821.2
|
Axin2
|
axin 2 |
| chr17_-_37938000 | 0.75 |
ENSMUST00000223366.2
ENSMUST00000216128.2 |
Olfr115
Olfr116
|
olfactory receptor 115 olfactory receptor 116 |
| chr12_+_9080014 | 0.74 |
ENSMUST00000219488.2
ENSMUST00000219470.2 |
Ttc32
|
tetratricopeptide repeat domain 32 |
| chr4_-_70453140 | 0.73 |
ENSMUST00000107359.9
|
Megf9
|
multiple EGF-like-domains 9 |
| chrM_+_7758 | 0.72 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr14_-_63415235 | 0.71 |
ENSMUST00000054963.10
|
Fdft1
|
farnesyl diphosphate farnesyl transferase 1 |
| chr19_+_46611826 | 0.71 |
ENSMUST00000111855.5
|
Wbp1l
|
WW domain binding protein 1 like |
| chr11_+_21041291 | 0.71 |
ENSMUST00000093290.12
|
Peli1
|
pellino 1 |
| chr2_-_26012751 | 0.70 |
ENSMUST00000140993.2
ENSMUST00000028300.6 |
Nacc2
|
nucleus accumbens associated 2, BEN and BTB (POZ) domain containing |
| chr4_-_15945359 | 0.70 |
ENSMUST00000029877.9
|
Decr1
|
2,4-dienoyl CoA reductase 1, mitochondrial |
| chrM_+_2743 | 0.69 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr6_+_68518603 | 0.69 |
ENSMUST00000168090.3
ENSMUST00000103326.3 |
Igkv1-99
|
immunoglobulin kappa variable 1-99 |
| chr13_-_21726945 | 0.68 |
ENSMUST00000205976.3
ENSMUST00000175637.3 |
Olfr1366
|
olfactory receptor 1366 |
| chr1_-_155022501 | 0.68 |
ENSMUST00000027744.10
|
Mr1
|
major histocompatibility complex, class I-related |
| chr4_-_114991478 | 0.66 |
ENSMUST00000106545.8
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr15_-_82872073 | 0.65 |
ENSMUST00000229439.2
|
Tcf20
|
transcription factor 20 |
| chr10_+_34359513 | 0.64 |
ENSMUST00000170771.3
|
Frk
|
fyn-related kinase |
| chr17_+_46807637 | 0.64 |
ENSMUST00000046497.8
|
Dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr13_-_14237980 | 0.64 |
ENSMUST00000222687.2
ENSMUST00000221338.2 ENSMUST00000221713.2 ENSMUST00000170957.3 |
Ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr10_+_28544556 | 0.63 |
ENSMUST00000161345.2
|
Themis
|
thymocyte selection associated |
| chr17_+_29487881 | 0.63 |
ENSMUST00000234845.2
ENSMUST00000235038.2 ENSMUST00000235050.2 ENSMUST00000120346.9 ENSMUST00000234377.2 ENSMUST00000235074.2 ENSMUST00000235040.2 ENSMUST00000234256.2 ENSMUST00000234459.2 |
BC004004
|
cDNA sequence BC004004 |
| chr1_-_130867810 | 0.63 |
ENSMUST00000112465.2
ENSMUST00000187410.7 ENSMUST00000187916.7 |
Il19
|
interleukin 19 |
| chr11_+_108811168 | 0.63 |
ENSMUST00000052915.14
|
Axin2
|
axin 2 |
| chr4_-_114991174 | 0.62 |
ENSMUST00000051400.8
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr1_+_191449946 | 0.62 |
ENSMUST00000133076.7
ENSMUST00000110855.8 |
Lpgat1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr16_-_37474772 | 0.61 |
ENSMUST00000023514.4
|
Ndufb4
|
NADH:ubiquinone oxidoreductase subunit B4 |
| chr18_+_13139992 | 0.60 |
ENSMUST00000041676.3
ENSMUST00000234084.2 ENSMUST00000234565.2 |
Hrh4
|
histamine receptor H4 |
| chr11_-_99276868 | 0.60 |
ENSMUST00000211768.2
|
Krt10
|
keratin 10 |
| chr14_+_31058589 | 0.60 |
ENSMUST00000022451.14
|
Capn7
|
calpain 7 |
| chr7_+_106740521 | 0.59 |
ENSMUST00000210474.2
|
Olfr716
|
olfactory receptor 716 |
| chr18_+_60426444 | 0.59 |
ENSMUST00000171297.2
|
F830016B08Rik
|
RIKEN cDNA F830016B08 gene |
| chr16_-_59459745 | 0.58 |
ENSMUST00000099646.10
|
Arl6
|
ADP-ribosylation factor-like 6 |
| chr18_+_37106851 | 0.58 |
ENSMUST00000192631.2
|
Pcdha7
|
protocadherin alpha 7 |
| chr12_-_56583582 | 0.58 |
ENSMUST00000001536.9
|
Nkx2-1
|
NK2 homeobox 1 |
| chr6_+_41107047 | 0.57 |
ENSMUST00000103271.2
|
Trbv13-3
|
T cell receptor beta, variable 13-3 |
| chr15_+_25742397 | 0.55 |
ENSMUST00000135981.8
|
Myo10
|
myosin X |
| chr2_+_30282266 | 0.55 |
ENSMUST00000028209.15
|
Dolpp1
|
dolichyl pyrophosphate phosphatase 1 |
| chr13_+_67052978 | 0.55 |
ENSMUST00000168767.9
|
Gm10767
|
predicted gene 10767 |
| chr10_-_117074501 | 0.54 |
ENSMUST00000159193.8
ENSMUST00000020392.5 |
9530003J23Rik
|
RIKEN cDNA 9530003J23 gene |
| chr1_-_183766195 | 0.54 |
ENSMUST00000050306.8
|
1700056E22Rik
|
RIKEN cDNA 1700056E22 gene |
| chr12_+_87840841 | 0.53 |
ENSMUST00000110147.3
|
Eif1ad3
|
eukaryotic translation initiation factor 1A domain containing 3 |
| chr1_+_177819001 | 0.53 |
ENSMUST00000094273.10
ENSMUST00000238506.2 ENSMUST00000239155.2 |
Catspere2
|
cation channel sperm associated auxiliary subunit epsilon 2 |
| chr8_-_45863572 | 0.52 |
ENSMUST00000209651.2
ENSMUST00000211370.2 ENSMUST00000034056.12 ENSMUST00000167106.3 |
Tlr3
|
toll-like receptor 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 18.5 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 2.5 | 7.4 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 2.3 | 6.9 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 1.8 | 21.6 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 1.5 | 6.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 1.4 | 21.0 | GO:0015747 | urate transport(GO:0015747) |
| 1.2 | 3.6 | GO:1904437 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.9 | 7.3 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.8 | 2.4 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.7 | 2.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.7 | 2.1 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.7 | 2.8 | GO:0061402 | glycerol biosynthetic process(GO:0006114) positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.7 | 3.4 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.7 | 2.0 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.6 | 6.2 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.6 | 1.8 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.6 | 3.6 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.6 | 4.0 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.6 | 2.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.5 | 2.2 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.5 | 3.7 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.5 | 5.5 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.5 | 2.9 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.4 | 1.8 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.4 | 1.7 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.4 | 1.6 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.4 | 1.1 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.3 | 1.0 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.3 | 1.3 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.3 | 3.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.3 | 0.9 | GO:0002780 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) |
| 0.3 | 3.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.3 | 1.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.3 | 1.5 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.3 | 1.3 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.2 | 6.9 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.2 | 0.7 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.2 | 2.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.2 | 3.0 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.2 | 0.7 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.2 | 1.4 | GO:2000054 | regulation of mismatch repair(GO:0032423) regulation of chondrocyte development(GO:0061181) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.2 | 2.5 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.2 | 1.2 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.2 | 0.8 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.2 | 0.6 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.2 | 1.3 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.2 | 2.8 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.2 | 1.8 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.2 | 1.6 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.2 | 1.4 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.2 | 0.8 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.2 | 1.1 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.2 | 5.0 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.2 | 2.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 2.0 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.4 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.1 | 0.8 | GO:0045356 | regulation of dendritic cell cytokine production(GO:0002730) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.1 | 1.4 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.1 | 0.6 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 0.3 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.1 | 1.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.3 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.1 | 1.2 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 1.0 | GO:0015961 | diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.1 | 0.3 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.1 | 0.4 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.1 | 4.0 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 0.3 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 0.4 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 0.3 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 0.3 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.2 | GO:1904000 | positive regulation of eating behavior(GO:1904000) |
| 0.1 | 0.4 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.1 | 0.2 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 0.5 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.1 | 0.6 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 0.1 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.1 | 2.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.5 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.1 | 0.3 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 0.4 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 | 0.9 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 1.0 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 0.7 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 1.3 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 0.6 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.1 | 0.3 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 1.1 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 0.6 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.2 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.1 | 0.4 | GO:0060266 | regulation of respiratory burst involved in inflammatory response(GO:0060264) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.1 | 0.3 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.9 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 0.1 | 0.3 | GO:0050758 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 9.3 | GO:0016052 | carbohydrate catabolic process(GO:0016052) |
| 0.1 | 0.3 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.1 | 0.9 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.1 | 0.5 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.1 | 1.6 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 1.1 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 2.6 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 7.0 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 2.5 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 3.7 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 1.8 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.2 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.0 | 1.8 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.9 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.9 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 2.9 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.4 | GO:0019682 | glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 | 0.7 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 2.1 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.6 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.0 | 3.6 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 0.5 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.5 | GO:0006577 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 0.0 | 0.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 1.0 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.9 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.3 | GO:0086028 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.0 | 0.6 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.7 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.2 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.2 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 2.9 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.0 | 1.4 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.7 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.2 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.6 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.8 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.6 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.4 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.5 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.9 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.3 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.6 | GO:0009988 | cell-cell recognition(GO:0009988) |
| 0.0 | 0.6 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 7.2 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 1.3 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.1 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.2 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 0.4 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.4 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.6 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.3 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 1.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.6 | GO:0001562 | response to protozoan(GO:0001562) |
| 0.0 | 0.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.2 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 1.1 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.1 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.0 | 0.3 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.0 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.8 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.5 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.0 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.0 | 0.3 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.5 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.4 | 3.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.4 | 1.1 | GO:0044317 | rod spherule(GO:0044317) |
| 0.3 | 18.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.3 | 4.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.3 | 1.3 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.2 | 2.6 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.2 | 7.9 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 2.8 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.2 | 2.3 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 0.7 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.2 | 0.5 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.2 | 3.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 2.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.4 | GO:0097632 | extrinsic component of pre-autophagosomal structure membrane(GO:0097632) |
| 0.1 | 13.0 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 1.6 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 3.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 2.6 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 1.1 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 0.3 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.1 | 0.2 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 1.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 4.3 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.5 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 2.5 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 2.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 9.5 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.4 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 3.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.1 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.4 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.3 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 0.3 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 10.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 6.2 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.4 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 6.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 5.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 33.7 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.2 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.9 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 3.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.5 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.5 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 20.6 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 3.1 | 18.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.4 | 10.0 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 1.4 | 5.5 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 1.2 | 3.7 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 1.2 | 21.0 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 1.1 | 7.6 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 1.0 | 6.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.9 | 5.6 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.9 | 2.8 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.8 | 3.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.7 | 6.4 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.6 | 1.7 | GO:0030338 | CMP-N-acetylneuraminate monooxygenase activity(GO:0030338) |
| 0.6 | 2.2 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.5 | 7.4 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.5 | 2.0 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.5 | 1.4 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.5 | 3.8 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.5 | 2.9 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.4 | 1.6 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.4 | 1.1 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.4 | 11.0 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 2.4 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.3 | 1.0 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.3 | 1.0 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.3 | 0.9 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.3 | 2.1 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.3 | 3.1 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.2 | 1.0 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.2 | 3.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.2 | 4.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 1.4 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.2 | 0.5 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.2 | 6.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 3.7 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.2 | 5.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.2 | 0.5 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.2 | 3.0 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.2 | 0.5 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.2 | 0.5 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.2 | 1.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.2 | 1.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.2 | 3.0 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 0.7 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.1 | 1.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.5 | GO:0047874 | dolichyldiphosphatase activity(GO:0047874) |
| 0.1 | 2.1 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.1 | 1.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.4 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.1 | 1.5 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.1 | 1.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 1.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 2.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.8 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 2.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 2.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.6 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.2 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 0.1 | 0.5 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 1.3 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.1 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.3 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 0.4 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.9 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 2.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 1.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 1.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.9 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 11.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 5.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.7 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 5.7 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 1.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.3 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.4 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 1.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.2 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.6 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.4 | GO:0004549 | tRNA-specific ribonuclease activity(GO:0004549) |
| 0.0 | 0.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 1.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 1.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.4 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.5 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 1.0 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 1.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.0 | 1.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.9 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 0.2 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.7 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.8 | GO:0004407 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.0 | 0.2 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 1.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.6 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 2.2 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.3 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 7.1 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 3.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 3.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 10.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 3.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.6 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.6 | 8.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.5 | 7.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.4 | 9.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 2.6 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.2 | 3.6 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 18.5 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.2 | 2.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 2.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 4.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.3 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 2.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 2.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 0.8 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 1.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 2.9 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 2.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 3.1 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 4.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 3.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.4 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.9 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 1.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.6 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 2.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |