Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
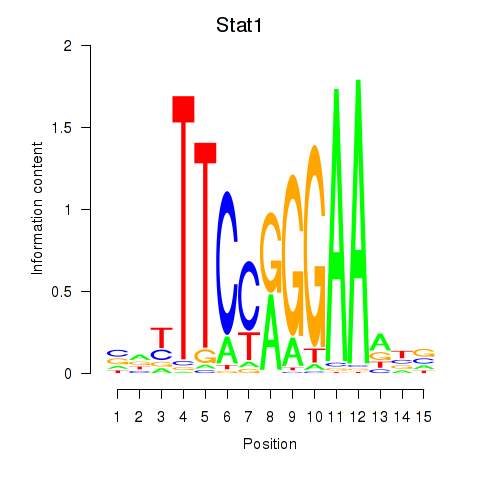
Results for Stat1
Z-value: 1.40
Transcription factors associated with Stat1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Stat1
|
ENSMUSG00000026104.15 | Stat1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Stat1 | mm39_v1_chr1_+_52158599_52158660 | 0.29 | 8.6e-02 | Click! |
Activity profile of Stat1 motif
Sorted Z-values of Stat1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Stat1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_34389269 | 10.46 |
ENSMUST00000007449.9
|
Akr1b7
|
aldo-keto reductase family 1, member B7 |
| chr9_+_110867807 | 9.76 |
ENSMUST00000197575.2
|
Ltf
|
lactotransferrin |
| chr4_+_114916703 | 5.61 |
ENSMUST00000162489.2
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr1_+_134110142 | 5.35 |
ENSMUST00000082060.10
ENSMUST00000153856.8 ENSMUST00000133701.8 ENSMUST00000132873.8 |
Chil1
|
chitinase-like 1 |
| chrX_-_8011952 | 5.26 |
ENSMUST00000115615.9
ENSMUST00000115616.8 ENSMUST00000115621.9 |
Rbm3
|
RNA binding motif (RNP1, RRM) protein 3 |
| chr4_+_120523758 | 5.07 |
ENSMUST00000094814.6
|
Cited4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr3_-_107145968 | 4.96 |
ENSMUST00000197758.5
|
Prok1
|
prokineticin 1 |
| chrX_-_8011918 | 4.90 |
ENSMUST00000115619.8
ENSMUST00000115617.10 ENSMUST00000040010.10 |
Rbm3
|
RNA binding motif (RNP1, RRM) protein 3 |
| chr14_-_70864448 | 4.31 |
ENSMUST00000110984.4
|
Dmtn
|
dematin actin binding protein |
| chr7_+_43086432 | 4.00 |
ENSMUST00000070518.4
|
Nkg7
|
natural killer cell group 7 sequence |
| chr1_+_134109888 | 3.70 |
ENSMUST00000156873.8
|
Chil1
|
chitinase-like 1 |
| chr6_+_41498716 | 3.47 |
ENSMUST00000070380.5
|
Prss2
|
protease, serine 2 |
| chr14_-_70864666 | 3.41 |
ENSMUST00000022694.17
|
Dmtn
|
dematin actin binding protein |
| chr6_+_40941688 | 3.18 |
ENSMUST00000076638.7
|
1810009J06Rik
|
RIKEN cDNA 1810009J06 gene |
| chr8_-_71060911 | 3.12 |
ENSMUST00000210580.2
ENSMUST00000211608.2 ENSMUST00000049908.11 |
Ssbp4
|
single stranded DNA binding protein 4 |
| chr7_+_43086554 | 3.04 |
ENSMUST00000206741.2
|
Nkg7
|
natural killer cell group 7 sequence |
| chrX_-_72974357 | 2.49 |
ENSMUST00000155597.2
ENSMUST00000114379.8 |
Renbp
|
renin binding protein |
| chr4_-_63965161 | 2.49 |
ENSMUST00000107377.10
|
Tnc
|
tenascin C |
| chr14_-_55950939 | 2.46 |
ENSMUST00000168729.8
ENSMUST00000228123.2 ENSMUST00000178034.9 |
Tgm1
|
transglutaminase 1, K polypeptide |
| chr3_+_96939732 | 2.41 |
ENSMUST00000132256.8
ENSMUST00000072600.7 |
Gja5
|
gap junction protein, alpha 5 |
| chr17_+_29309942 | 2.40 |
ENSMUST00000119901.9
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chr5_-_140687995 | 2.36 |
ENSMUST00000135028.5
ENSMUST00000077890.12 ENSMUST00000041783.14 ENSMUST00000142081.6 |
Iqce
|
IQ motif containing E |
| chrX_-_72974440 | 2.22 |
ENSMUST00000116578.8
|
Renbp
|
renin binding protein |
| chr15_-_97729341 | 1.97 |
ENSMUST00000079838.14
ENSMUST00000118294.8 |
Hdac7
|
histone deacetylase 7 |
| chr11_+_69886603 | 1.87 |
ENSMUST00000018716.10
|
Phf23
|
PHD finger protein 23 |
| chr7_-_99770653 | 1.86 |
ENSMUST00000208670.2
ENSMUST00000032969.14 |
Pold3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr6_+_129374260 | 1.84 |
ENSMUST00000032262.14
|
Clec1b
|
C-type lectin domain family 1, member b |
| chr8_+_121463090 | 1.84 |
ENSMUST00000160943.3
ENSMUST00000047737.10 ENSMUST00000162658.8 |
Irf8
|
interferon regulatory factor 8 |
| chr3_-_96201248 | 1.77 |
ENSMUST00000029748.8
|
Fcgr1
|
Fc receptor, IgG, high affinity I |
| chr11_+_69886652 | 1.73 |
ENSMUST00000101526.9
|
Phf23
|
PHD finger protein 23 |
| chr10_-_61619790 | 1.69 |
ENSMUST00000020283.5
|
Macroh2a2
|
macroH2A.2 histone |
| chr3_-_144412394 | 1.63 |
ENSMUST00000200532.2
|
Sh3glb1
|
SH3-domain GRB2-like B1 (endophilin) |
| chr4_+_114945905 | 1.59 |
ENSMUST00000171877.8
ENSMUST00000177647.8 ENSMUST00000106548.9 ENSMUST00000030488.3 |
Pdzk1ip1
|
PDZK1 interacting protein 1 |
| chr6_+_129374441 | 1.54 |
ENSMUST00000112081.9
ENSMUST00000112079.3 |
Clec1b
|
C-type lectin domain family 1, member b |
| chr2_-_163592127 | 1.44 |
ENSMUST00000017841.4
|
Ada
|
adenosine deaminase |
| chr5_+_136023649 | 1.39 |
ENSMUST00000111142.9
ENSMUST00000111145.10 ENSMUST00000111144.8 ENSMUST00000199239.5 ENSMUST00000005072.10 ENSMUST00000130345.2 |
Dtx2
|
deltex 2, E3 ubiquitin ligase |
| chr3_-_88417251 | 1.39 |
ENSMUST00000149068.2
|
Lmna
|
lamin A |
| chr7_-_99770280 | 1.30 |
ENSMUST00000208184.2
|
Pold3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr12_-_91351177 | 1.30 |
ENSMUST00000141429.8
|
Cep128
|
centrosomal protein 128 |
| chr6_+_42326760 | 1.26 |
ENSMUST00000203652.3
ENSMUST00000070635.13 |
Zyx
|
zyxin |
| chr4_+_63478478 | 1.24 |
ENSMUST00000080336.4
|
Tmem268
|
transmembrane protein 268 |
| chr19_-_46950355 | 1.23 |
ENSMUST00000236501.2
|
Nt5c2
|
5'-nucleotidase, cytosolic II |
| chr7_+_79836581 | 1.23 |
ENSMUST00000032754.9
|
Sema4b
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr16_-_20245071 | 1.21 |
ENSMUST00000115547.9
ENSMUST00000096199.5 |
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr6_+_42326714 | 1.18 |
ENSMUST00000203846.3
|
Zyx
|
zyxin |
| chr10_+_128139227 | 1.13 |
ENSMUST00000218315.2
ENSMUST00000219721.2 |
Pan2
|
PAN2 poly(A) specific ribonuclease subunit |
| chr10_-_95252712 | 1.13 |
ENSMUST00000020215.16
|
Socs2
|
suppressor of cytokine signaling 2 |
| chr7_-_30643444 | 1.12 |
ENSMUST00000062620.9
|
Hamp
|
hepcidin antimicrobial peptide |
| chr4_+_63478454 | 1.09 |
ENSMUST00000124332.8
ENSMUST00000150360.8 |
Tmem268
|
transmembrane protein 268 |
| chr9_-_30833748 | 1.08 |
ENSMUST00000065112.7
|
Adamts15
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 15 |
| chr7_-_127185512 | 1.05 |
ENSMUST00000205839.2
|
Ccdc189
|
coiled-coil domain containing 189 |
| chr9_+_86625694 | 1.05 |
ENSMUST00000179574.2
ENSMUST00000036426.13 |
Prss35
|
protease, serine 35 |
| chr12_-_112637998 | 1.04 |
ENSMUST00000128300.9
|
Akt1
|
thymoma viral proto-oncogene 1 |
| chr1_+_171594690 | 1.01 |
ENSMUST00000015460.5
|
Slamf1
|
signaling lymphocytic activation molecule family member 1 |
| chr15_-_93417380 | 1.00 |
ENSMUST00000109255.3
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chr5_-_148988110 | 1.00 |
ENSMUST00000110505.8
|
Hmgb1
|
high mobility group box 1 |
| chr4_-_129472328 | 1.00 |
ENSMUST00000052835.9
|
Fam167b
|
family with sequence similarity 167, member B |
| chr10_+_128139191 | 1.00 |
ENSMUST00000005825.8
|
Pan2
|
PAN2 poly(A) specific ribonuclease subunit |
| chr11_+_101136821 | 1.00 |
ENSMUST00000129680.8
|
Ramp2
|
receptor (calcitonin) activity modifying protein 2 |
| chr16_-_20245138 | 0.96 |
ENSMUST00000079158.13
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr11_-_59937302 | 0.96 |
ENSMUST00000000310.14
ENSMUST00000102693.9 ENSMUST00000148512.2 |
Pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr15_-_37459570 | 0.95 |
ENSMUST00000119730.8
ENSMUST00000120746.8 |
Ncald
|
neurocalcin delta |
| chr4_-_136613498 | 0.92 |
ENSMUST00000046384.9
|
C1qb
|
complement component 1, q subcomponent, beta polypeptide |
| chr7_-_100232276 | 0.91 |
ENSMUST00000152876.3
ENSMUST00000150042.8 ENSMUST00000132888.9 |
Mrpl48
|
mitochondrial ribosomal protein L48 |
| chr2_+_150590956 | 0.90 |
ENSMUST00000094467.6
|
Entpd6
|
ectonucleoside triphosphate diphosphohydrolase 6 |
| chr6_-_65121892 | 0.89 |
ENSMUST00000031982.5
|
Hpgds
|
hematopoietic prostaglandin D synthase |
| chr6_+_28475099 | 0.88 |
ENSMUST00000168362.2
|
Snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr5_+_90708962 | 0.83 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr7_-_126224848 | 0.79 |
ENSMUST00000032961.4
|
Nupr1
|
nuclear protein transcription regulator 1 |
| chr6_+_42326528 | 0.78 |
ENSMUST00000203329.3
|
Zyx
|
zyxin |
| chr11_-_45846291 | 0.78 |
ENSMUST00000011398.13
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr8_+_11778039 | 0.75 |
ENSMUST00000110909.9
|
Arhgef7
|
Rho guanine nucleotide exchange factor (GEF7) |
| chr6_-_122317484 | 0.71 |
ENSMUST00000112600.9
|
Phc1
|
polyhomeotic 1 |
| chr7_-_44665639 | 0.69 |
ENSMUST00000085383.11
|
Scaf1
|
SR-related CTD-associated factor 1 |
| chr4_-_129534853 | 0.67 |
ENSMUST00000046425.16
ENSMUST00000133803.8 |
Txlna
|
taxilin alpha |
| chr4_-_129534752 | 0.66 |
ENSMUST00000132217.8
ENSMUST00000130017.2 ENSMUST00000154105.8 |
Txlna
|
taxilin alpha |
| chr7_-_100505486 | 0.65 |
ENSMUST00000139604.8
|
Relt
|
RELT tumor necrosis factor receptor |
| chr12_+_86129329 | 0.65 |
ENSMUST00000054565.8
ENSMUST00000222821.2 ENSMUST00000222905.2 |
Ift43
|
intraflagellar transport 43 |
| chr19_+_37184927 | 0.64 |
ENSMUST00000024078.15
ENSMUST00000112391.8 |
Marchf5
|
membrane associated ring-CH-type finger 5 |
| chr7_-_108774367 | 0.63 |
ENSMUST00000207178.2
|
Lmo1
|
LIM domain only 1 |
| chr8_+_11777721 | 0.63 |
ENSMUST00000210104.2
|
Arhgef7
|
Rho guanine nucleotide exchange factor (GEF7) |
| chr17_-_48145466 | 0.62 |
ENSMUST00000066368.13
|
Mdfi
|
MyoD family inhibitor |
| chr8_+_71126012 | 0.62 |
ENSMUST00000146972.3
ENSMUST00000210071.2 ENSMUST00000210987.2 |
Lsm4
|
LSM4 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr14_-_118943591 | 0.61 |
ENSMUST00000036554.14
ENSMUST00000166646.2 |
Abcc4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr17_+_36134398 | 0.61 |
ENSMUST00000173493.8
ENSMUST00000173147.8 |
Flot1
|
flotillin 1 |
| chr18_-_33596890 | 0.61 |
ENSMUST00000237066.2
|
Nrep
|
neuronal regeneration related protein |
| chr6_-_48025845 | 0.61 |
ENSMUST00000095944.10
|
Zfp777
|
zinc finger protein 777 |
| chr14_+_55842002 | 0.60 |
ENSMUST00000138037.2
|
Irf9
|
interferon regulatory factor 9 |
| chr18_-_38417390 | 0.59 |
ENSMUST00000025311.8
|
Pcdh12
|
protocadherin 12 |
| chr8_-_112064783 | 0.57 |
ENSMUST00000056157.14
ENSMUST00000120432.3 |
Mlkl
|
mixed lineage kinase domain-like |
| chr10_-_78427721 | 0.57 |
ENSMUST00000040580.7
|
Syde1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans) |
| chr8_-_125296435 | 0.56 |
ENSMUST00000238882.2
ENSMUST00000063278.7 |
Agt
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8) |
| chr8_+_71125876 | 0.55 |
ENSMUST00000034311.15
|
Lsm4
|
LSM4 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr3_+_89136353 | 0.54 |
ENSMUST00000041142.4
|
Muc1
|
mucin 1, transmembrane |
| chr11_-_98620200 | 0.54 |
ENSMUST00000126565.2
ENSMUST00000100500.9 ENSMUST00000017354.13 |
Med24
|
mediator complex subunit 24 |
| chr3_-_108117754 | 0.52 |
ENSMUST00000117784.8
ENSMUST00000119650.8 ENSMUST00000117409.8 |
Atxn7l2
|
ataxin 7-like 2 |
| chr2_+_128942900 | 0.52 |
ENSMUST00000103205.11
|
Polr1b
|
polymerase (RNA) I polypeptide B |
| chr2_+_128942919 | 0.52 |
ENSMUST00000028874.8
|
Polr1b
|
polymerase (RNA) I polypeptide B |
| chr10_-_79911245 | 0.50 |
ENSMUST00000217972.2
|
Sbno2
|
strawberry notch 2 |
| chr10_-_81102740 | 0.50 |
ENSMUST00000046114.5
|
Mrpl54
|
mitochondrial ribosomal protein L54 |
| chr10_+_40225272 | 0.48 |
ENSMUST00000044672.11
ENSMUST00000095743.4 |
Cdk19
|
cyclin-dependent kinase 19 |
| chr18_+_42644552 | 0.48 |
ENSMUST00000237602.2
ENSMUST00000236088.2 ENSMUST00000025375.15 |
Tcerg1
|
transcription elongation regulator 1 (CA150) |
| chr4_-_129534403 | 0.47 |
ENSMUST00000084264.12
|
Txlna
|
taxilin alpha |
| chr5_+_75312939 | 0.46 |
ENSMUST00000202681.4
ENSMUST00000000476.15 |
Pdgfra
|
platelet derived growth factor receptor, alpha polypeptide |
| chr10_-_95253042 | 0.45 |
ENSMUST00000135822.8
|
Socs2
|
suppressor of cytokine signaling 2 |
| chr2_-_126341757 | 0.45 |
ENSMUST00000040128.12
|
Atp8b4
|
ATPase, class I, type 8B, member 4 |
| chrX_+_158086253 | 0.44 |
ENSMUST00000112491.2
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr17_+_36134122 | 0.44 |
ENSMUST00000001569.15
ENSMUST00000174080.8 |
Flot1
|
flotillin 1 |
| chr9_+_86454018 | 0.44 |
ENSMUST00000185566.7
ENSMUST00000034988.10 ENSMUST00000179212.3 |
Rwdd2a
|
RWD domain containing 2A |
| chr1_+_171041539 | 0.44 |
ENSMUST00000005820.11
ENSMUST00000075469.12 ENSMUST00000155126.8 |
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr18_-_33596468 | 0.43 |
ENSMUST00000171533.9
|
Nrep
|
neuronal regeneration related protein |
| chr6_+_127423779 | 0.43 |
ENSMUST00000112191.8
|
Parp11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr18_-_52662917 | 0.43 |
ENSMUST00000171470.8
|
Lox
|
lysyl oxidase |
| chr13_-_46881388 | 0.40 |
ENSMUST00000021803.10
|
Nup153
|
nucleoporin 153 |
| chr18_-_38417444 | 0.39 |
ENSMUST00000194012.2
|
Pcdh12
|
protocadherin 12 |
| chr5_+_52991351 | 0.38 |
ENSMUST00000031072.14
|
Anapc4
|
anaphase promoting complex subunit 4 |
| chr1_+_36346824 | 0.38 |
ENSMUST00000142319.8
ENSMUST00000097778.9 ENSMUST00000115031.8 ENSMUST00000115032.8 ENSMUST00000137906.2 ENSMUST00000115029.2 |
Arid5a
|
AT rich interactive domain 5A (MRF1-like) |
| chr16_-_31898088 | 0.38 |
ENSMUST00000023467.9
|
Pak2
|
p21 (RAC1) activated kinase 2 |
| chr19_-_46315543 | 0.37 |
ENSMUST00000223917.2
ENSMUST00000224447.2 ENSMUST00000041391.5 ENSMUST00000096029.12 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr16_+_17326810 | 0.37 |
ENSMUST00000231292.2
|
Lztr1
|
leucine-zipper-like transcriptional regulator, 1 |
| chr18_-_33596792 | 0.37 |
ENSMUST00000051087.16
|
Nrep
|
neuronal regeneration related protein |
| chr1_+_171041583 | 0.36 |
ENSMUST00000111328.8
|
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr19_-_40260286 | 0.36 |
ENSMUST00000182432.2
|
Pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr18_-_52662728 | 0.36 |
ENSMUST00000025409.9
|
Lox
|
lysyl oxidase |
| chr9_+_44893077 | 0.35 |
ENSMUST00000034602.9
|
Cd3d
|
CD3 antigen, delta polypeptide |
| chr16_+_17327076 | 0.35 |
ENSMUST00000232242.2
|
Lztr1
|
leucine-zipper-like transcriptional regulator, 1 |
| chr17_+_36134450 | 0.35 |
ENSMUST00000172846.2
|
Flot1
|
flotillin 1 |
| chr11_+_81926394 | 0.35 |
ENSMUST00000000193.6
|
Ccl2
|
chemokine (C-C motif) ligand 2 |
| chr7_+_45204317 | 0.34 |
ENSMUST00000107752.12
|
Hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr17_+_34364206 | 0.34 |
ENSMUST00000041982.9
ENSMUST00000171231.8 |
H2-DMb2
|
histocompatibility 2, class II, locus Mb2 |
| chr5_+_35198853 | 0.34 |
ENSMUST00000030985.10
ENSMUST00000202573.2 |
Hgfac
|
hepatocyte growth factor activator |
| chr7_+_89780785 | 0.32 |
ENSMUST00000208684.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr8_-_106052884 | 0.30 |
ENSMUST00000210412.2
ENSMUST00000210801.2 ENSMUST00000070508.8 |
Lrrc29
|
leucine rich repeat containing 29 |
| chr10_+_20188207 | 0.30 |
ENSMUST00000092678.10
ENSMUST00000043881.12 |
Bclaf1
|
BCL2-associated transcription factor 1 |
| chr9_-_123507937 | 0.29 |
ENSMUST00000040960.13
|
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr9_-_71393175 | 0.27 |
ENSMUST00000233263.2
ENSMUST00000034720.12 |
Polr2m
|
polymerase (RNA) II (DNA directed) polypeptide M |
| chr19_-_40260060 | 0.27 |
ENSMUST00000068439.13
|
Pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr17_+_28910302 | 0.21 |
ENSMUST00000004990.14
ENSMUST00000114754.8 ENSMUST00000062694.16 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chr18_+_37453427 | 0.20 |
ENSMUST00000078271.4
|
Pcdhb5
|
protocadherin beta 5 |
| chr18_-_43610829 | 0.19 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr11_+_53660834 | 0.19 |
ENSMUST00000108920.10
ENSMUST00000140866.9 ENSMUST00000108922.9 |
Irf1
|
interferon regulatory factor 1 |
| chr17_+_28910393 | 0.18 |
ENSMUST00000124886.9
ENSMUST00000114758.9 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chr10_+_20188269 | 0.18 |
ENSMUST00000185800.7
|
Bclaf1
|
BCL2-associated transcription factor 1 |
| chrX_-_99670174 | 0.18 |
ENSMUST00000015812.12
|
Pdzd11
|
PDZ domain containing 11 |
| chr6_-_122317457 | 0.17 |
ENSMUST00000160843.8
|
Phc1
|
polyhomeotic 1 |
| chr5_-_34671236 | 0.17 |
ENSMUST00000114359.2
ENSMUST00000030991.14 ENSMUST00000087737.10 |
Tnip2
|
TNFAIP3 interacting protein 2 |
| chr16_-_20244631 | 0.16 |
ENSMUST00000077867.10
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr14_-_122153185 | 0.15 |
ENSMUST00000055475.9
|
Gpr18
|
G protein-coupled receptor 18 |
| chr9_-_44437694 | 0.15 |
ENSMUST00000062215.8
|
Cxcr5
|
chemokine (C-X-C motif) receptor 5 |
| chr8_-_112064393 | 0.15 |
ENSMUST00000145862.3
|
Mlkl
|
mixed lineage kinase domain-like |
| chr19_+_36811615 | 0.15 |
ENSMUST00000025729.12
|
Tnks2
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2 |
| chrX_-_8118541 | 0.14 |
ENSMUST00000115594.8
ENSMUST00000115595.8 ENSMUST00000033513.10 |
Ftsj1
|
FtsJ RNA methyltransferase homolog 1 (E. coli) |
| chr12_-_11315755 | 0.13 |
ENSMUST00000166117.4
ENSMUST00000219600.2 ENSMUST00000218487.2 |
Gen1
|
GEN1, Holliday junction 5' flap endonuclease |
| chr11_-_72106418 | 0.13 |
ENSMUST00000021157.9
|
Med31
|
mediator complex subunit 31 |
| chr5_-_6926523 | 0.12 |
ENSMUST00000164784.2
|
Zfp804b
|
zinc finger protein 804B |
| chr4_+_83335947 | 0.12 |
ENSMUST00000030206.10
ENSMUST00000071544.11 |
Snapc3
|
small nuclear RNA activating complex, polypeptide 3 |
| chr16_+_21613068 | 0.11 |
ENSMUST00000211443.2
ENSMUST00000231300.2 ENSMUST00000209449.2 ENSMUST00000181780.9 ENSMUST00000209728.2 ENSMUST00000181960.3 ENSMUST00000209429.2 ENSMUST00000180830.3 ENSMUST00000231988.2 |
1300002E11Rik
Map3k13
|
RIKEN cDNA 1300002E11 gene mitogen-activated protein kinase kinase kinase 13 |
| chr15_-_37458768 | 0.10 |
ENSMUST00000116445.9
|
Ncald
|
neurocalcin delta |
| chr17_+_34406523 | 0.09 |
ENSMUST00000170086.8
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr16_-_42160957 | 0.08 |
ENSMUST00000102817.5
|
Gap43
|
growth associated protein 43 |
| chr11_+_101315893 | 0.08 |
ENSMUST00000040561.6
|
Rundc1
|
RUN domain containing 1 |
| chr15_+_34238174 | 0.07 |
ENSMUST00000022867.5
ENSMUST00000226627.2 |
Laptm4b
|
lysosomal-associated protein transmembrane 4B |
| chr16_+_91169770 | 0.06 |
ENSMUST00000089042.7
|
Ifnar2
|
interferon (alpha and beta) receptor 2 |
| chr11_+_96173355 | 0.05 |
ENSMUST00000125410.2
|
Hoxb8
|
homeobox B8 |
| chr17_+_34406762 | 0.05 |
ENSMUST00000041633.15
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr8_-_71229293 | 0.05 |
ENSMUST00000034296.15
|
Pik3r2
|
phosphoinositide-3-kinase regulatory subunit 2 |
| chr16_-_59138611 | 0.04 |
ENSMUST00000216261.2
|
Olfr204
|
olfactory receptor 204 |
| chr2_-_117173312 | 0.04 |
ENSMUST00000178884.8
|
Rasgrp1
|
RAS guanyl releasing protein 1 |
| chr2_-_117173190 | 0.03 |
ENSMUST00000173541.8
ENSMUST00000172901.8 ENSMUST00000173252.2 |
Rasgrp1
|
RAS guanyl releasing protein 1 |
| chr2_-_37312881 | 0.03 |
ENSMUST00000112936.4
ENSMUST00000112934.8 |
Rc3h2
|
ring finger and CCCH-type zinc finger domains 2 |
| chr12_-_108859123 | 0.02 |
ENSMUST00000161154.2
ENSMUST00000161410.8 |
Wars
|
tryptophanyl-tRNA synthetase |
| chr8_+_26022141 | 0.02 |
ENSMUST00000210846.2
ENSMUST00000167764.2 |
Fgfr1
|
fibroblast growth factor receptor 1 |
| chr3_+_138233004 | 0.02 |
ENSMUST00000196990.5
ENSMUST00000200239.5 ENSMUST00000200100.2 |
Eif4e
|
eukaryotic translation initiation factor 4E |
| chr2_+_101716577 | 0.01 |
ENSMUST00000028584.8
|
Commd9
|
COMM domain containing 9 |
| chr3_-_57202301 | 0.01 |
ENSMUST00000171384.8
|
Tm4sf1
|
transmembrane 4 superfamily member 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.8 | GO:0044010 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 1.9 | 5.6 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 1.3 | 7.7 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.8 | 2.4 | GO:0010645 | atrial ventricular junction remodeling(GO:0003294) regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.7 | 4.7 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.6 | 2.5 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.6 | 1.8 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.5 | 1.6 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.5 | 1.4 | GO:0002314 | germinal center B cell differentiation(GO:0002314) adenosine catabolic process(GO:0006154) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) purine deoxyribonucleoside metabolic process(GO:0046122) negative regulation of mucus secretion(GO:0070256) |
| 0.4 | 3.6 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.4 | 9.6 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.4 | 3.2 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.3 | 1.0 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.3 | 1.0 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.3 | 1.0 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.3 | 1.1 | GO:0034760 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 0.3 | 1.0 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.3 | 1.0 | GO:0017126 | nucleologenesis(GO:0017126) |
| 0.2 | 1.4 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.2 | 1.4 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 2.5 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.2 | 10.2 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.2 | 3.5 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.2 | 0.5 | GO:0038091 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.2 | 2.4 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.1 | 0.9 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 1.0 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 | 0.4 | GO:0046832 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.1 | 0.5 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.1 | 1.0 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 2.0 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.3 | GO:0035702 | immune complex clearance by monocytes and macrophages(GO:0002436) monocyte homeostasis(GO:0035702) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) |
| 0.1 | 0.6 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 2.1 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.1 | 0.4 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.1 | 0.4 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 1.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.1 | 0.3 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.1 | 0.5 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 1.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 1.8 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.1 | 2.7 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 0.3 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.1 | 0.3 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 0.2 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.5 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 1.6 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.1 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.0 | 0.6 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.8 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.0 | 4.8 | GO:0043627 | response to estrogen(GO:0043627) |
| 0.0 | 1.2 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.2 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.0 | 0.4 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 0.6 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.6 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.4 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 1.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 4.6 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.3 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.1 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.7 | GO:0070266 | necroptotic process(GO:0070266) |
| 0.0 | 0.2 | GO:0023035 | CD40 signaling pathway(GO:0023035) toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 | 0.9 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.9 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 1.7 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.6 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 1.6 | 9.8 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 1.1 | 7.7 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.8 | 2.4 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.4 | 2.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 1.4 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 1.0 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.2 | 0.6 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 1.4 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 2.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 1.4 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 2.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.9 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 11.4 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.1 | 1.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.9 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 0.6 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 1.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.4 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.3 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.3 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 1.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 5.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.4 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 2.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.0 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 1.7 | GO:0000922 | spindle pole(GO:0000922) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 10.5 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 1.1 | 10.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.8 | 2.4 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.8 | 9.0 | GO:0008061 | chitin binding(GO:0008061) |
| 0.6 | 2.4 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.3 | 1.0 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.3 | 9.8 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.3 | 4.7 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.3 | 1.6 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.3 | 0.8 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.3 | 1.8 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.2 | 2.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 2.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 0.9 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.2 | 1.0 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.2 | 0.5 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.8 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 1.4 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 1.0 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 2.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 2.0 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.0 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 5.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 7.1 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 5.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 0.8 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 1.6 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.1 | 1.0 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.1 | 1.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.0 | 0.9 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.6 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.3 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.6 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 1.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 4.0 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.7 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 1.1 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 1.8 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.7 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 1.7 | GO:0043492 | ATPase activity, coupled to movement of substances(GO:0043492) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 3.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 0.6 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 2.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.8 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 1.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 4.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 3.5 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.2 | 3.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 2.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 2.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 2.9 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 1.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.0 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 2.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.4 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 5.6 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 1.9 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 1.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | Genes involved in Gastrin-CREB signalling pathway via PKC and MAPK |
| 0.0 | 0.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.3 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.4 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 1.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |