Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Tal1
Z-value: 7.19
Transcription factors associated with Tal1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tal1
|
ENSMUSG00000028717.13 | Tal1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tal1 | mm39_v1_chr4_+_114916703_114916759 | 0.97 | 9.3e-23 | Click! |
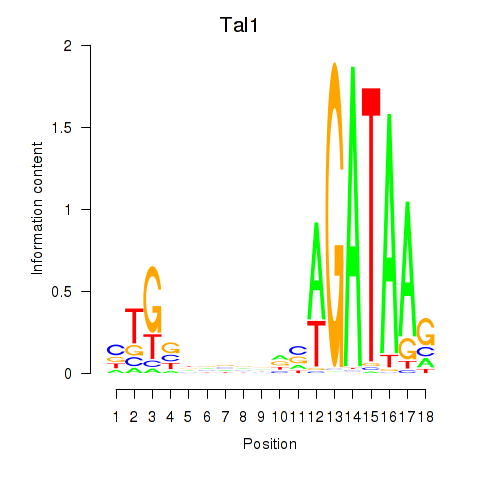
Activity profile of Tal1 motif
Sorted Z-values of Tal1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tal1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_103159892 | 90.35 |
ENSMUST00000133600.8
ENSMUST00000134554.2 ENSMUST00000156927.8 |
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr15_-_103160082 | 82.43 |
ENSMUST00000149111.8
ENSMUST00000132836.8 |
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr17_+_41121979 | 70.59 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr1_-_132295617 | 59.00 |
ENSMUST00000142609.8
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr17_+_37180437 | 56.90 |
ENSMUST00000060524.11
|
Trim10
|
tripartite motif-containing 10 |
| chr4_+_134591847 | 56.23 |
ENSMUST00000030627.8
|
Rhd
|
Rh blood group, D antigen |
| chr6_+_86055018 | 52.17 |
ENSMUST00000205034.3
ENSMUST00000203724.3 |
Add2
|
adducin 2 (beta) |
| chr4_-_119047202 | 51.84 |
ENSMUST00000239029.2
ENSMUST00000138395.9 ENSMUST00000156746.3 |
Ermap
|
erythroblast membrane-associated protein |
| chr8_+_81220410 | 46.28 |
ENSMUST00000063359.8
|
Gypa
|
glycophorin A |
| chr1_+_131566044 | 45.81 |
ENSMUST00000073350.13
|
Ctse
|
cathepsin E |
| chr17_-_26417982 | 45.61 |
ENSMUST00000142410.2
ENSMUST00000120333.8 ENSMUST00000039113.14 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr5_+_90920294 | 45.33 |
ENSMUST00000031320.8
|
Pf4
|
platelet factor 4 |
| chr14_-_44112974 | 44.63 |
ENSMUST00000179200.2
|
Ear1
|
eosinophil-associated, ribonuclease A family, member 1 |
| chr1_+_131566223 | 43.76 |
ENSMUST00000112411.2
|
Ctse
|
cathepsin E |
| chr8_+_85428059 | 43.12 |
ENSMUST00000238364.2
ENSMUST00000238562.2 ENSMUST00000037165.6 |
Lyl1
|
lymphoblastomic leukemia 1 |
| chr4_-_119047167 | 42.52 |
ENSMUST00000030396.15
|
Ermap
|
erythroblast membrane-associated protein |
| chr9_-_21874802 | 42.46 |
ENSMUST00000006397.7
|
Epor
|
erythropoietin receptor |
| chr6_+_86055048 | 42.00 |
ENSMUST00000032069.8
|
Add2
|
adducin 2 (beta) |
| chr5_+_90920353 | 41.47 |
ENSMUST00000202625.2
|
Pf4
|
platelet factor 4 |
| chrX_+_8137881 | 39.55 |
ENSMUST00000115590.2
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr7_-_103502404 | 36.52 |
ENSMUST00000033229.5
|
Hbb-y
|
hemoglobin Y, beta-like embryonic chain |
| chr1_+_174000304 | 36.46 |
ENSMUST00000027817.8
|
Spta1
|
spectrin alpha, erythrocytic 1 |
| chr14_-_44057096 | 36.33 |
ENSMUST00000100691.4
|
Ear1
|
eosinophil-associated, ribonuclease A family, member 1 |
| chr11_-_79971750 | 36.12 |
ENSMUST00000103233.10
ENSMUST00000061283.15 |
Crlf3
|
cytokine receptor-like factor 3 |
| chr10_-_62178453 | 35.31 |
ENSMUST00000143179.2
ENSMUST00000130422.8 |
Hk1
|
hexokinase 1 |
| chr4_-_119047180 | 33.51 |
ENSMUST00000150864.3
ENSMUST00000141227.9 |
Ermap
|
erythroblast membrane-associated protein |
| chr4_-_119047146 | 33.38 |
ENSMUST00000124626.9
|
Ermap
|
erythroblast membrane-associated protein |
| chr7_+_99243812 | 32.04 |
ENSMUST00000162290.2
|
Arrb1
|
arrestin, beta 1 |
| chr9_+_20940669 | 31.51 |
ENSMUST00000001040.7
ENSMUST00000215077.2 |
Icam4
|
intercellular adhesion molecule 4, Landsteiner-Wiener blood group |
| chr14_+_52091156 | 31.12 |
ENSMUST00000169070.2
ENSMUST00000074477.7 |
Ear6
|
eosinophil-associated, ribonuclease A family, member 6 |
| chr16_-_18440388 | 30.89 |
ENSMUST00000167388.3
|
Gp1bb
|
glycoprotein Ib, beta polypeptide |
| chr8_+_23525101 | 28.77 |
ENSMUST00000117662.8
ENSMUST00000117296.8 ENSMUST00000141784.9 |
Ank1
|
ankyrin 1, erythroid |
| chr17_+_31427023 | 26.79 |
ENSMUST00000173776.2
ENSMUST00000048656.15 |
Ubash3a
|
ubiquitin associated and SH3 domain containing, A |
| chr1_-_181669891 | 26.29 |
ENSMUST00000193028.2
ENSMUST00000191878.6 ENSMUST00000005003.12 |
Lbr
|
lamin B receptor |
| chr11_+_32226400 | 25.67 |
ENSMUST00000020531.9
|
Hba-x
|
hemoglobin X, alpha-like embryonic chain in Hba complex |
| chr6_+_41331039 | 24.75 |
ENSMUST00000072103.7
|
Try10
|
trypsin 10 |
| chr2_+_84669739 | 22.54 |
ENSMUST00000146816.8
ENSMUST00000028469.14 |
Slc43a1
|
solute carrier family 43, member 1 |
| chr15_+_80507671 | 22.36 |
ENSMUST00000043149.9
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr15_+_78810919 | 22.09 |
ENSMUST00000089377.6
|
Lgals1
|
lectin, galactose binding, soluble 1 |
| chr11_+_58808830 | 21.99 |
ENSMUST00000020792.12
ENSMUST00000108818.4 |
Btnl10
|
butyrophilin-like 10 |
| chr17_-_33932346 | 21.32 |
ENSMUST00000173392.2
|
Marchf2
|
membrane associated ring-CH-type finger 2 |
| chrX_-_106446928 | 21.11 |
ENSMUST00000033591.6
|
Itm2a
|
integral membrane protein 2A |
| chr6_-_136834725 | 21.04 |
ENSMUST00000032341.3
|
Art4
|
ADP-ribosyltransferase 4 |
| chr8_+_95712151 | 20.93 |
ENSMUST00000212799.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr11_-_102360664 | 20.34 |
ENSMUST00000103086.4
|
Itga2b
|
integrin alpha 2b |
| chr4_+_132701413 | 19.91 |
ENSMUST00000030693.13
|
Fgr
|
FGR proto-oncogene, Src family tyrosine kinase |
| chr12_+_111387978 | 19.66 |
ENSMUST00000222897.2
|
Exoc3l4
|
exocyst complex component 3-like 4 |
| chr7_+_110372860 | 19.65 |
ENSMUST00000143786.2
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr7_-_4400704 | 19.57 |
ENSMUST00000108590.4
ENSMUST00000206928.2 |
Gp6
|
glycoprotein 6 (platelet) |
| chr3_-_14873406 | 19.22 |
ENSMUST00000181860.8
ENSMUST00000144327.3 |
Car1
|
carbonic anhydrase 1 |
| chr8_-_86091946 | 19.18 |
ENSMUST00000034133.14
|
Mylk3
|
myosin light chain kinase 3 |
| chr11_+_32226893 | 19.16 |
ENSMUST00000145569.2
|
Hba-x
|
hemoglobin X, alpha-like embryonic chain in Hba complex |
| chr6_+_41435846 | 19.05 |
ENSMUST00000031910.8
|
Prss1
|
protease, serine 1 (trypsin 1) |
| chrX_+_92718695 | 18.02 |
ENSMUST00000045898.4
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr9_+_107828136 | 17.26 |
ENSMUST00000049348.9
ENSMUST00000194271.2 |
Traip
|
TRAF-interacting protein |
| chr14_+_26722319 | 17.18 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr17_+_35133435 | 16.76 |
ENSMUST00000007249.15
|
Slc44a4
|
solute carrier family 44, member 4 |
| chr8_+_85428391 | 16.50 |
ENSMUST00000238338.2
|
Lyl1
|
lymphoblastomic leukemia 1 |
| chr9_-_110886306 | 16.45 |
ENSMUST00000195968.2
ENSMUST00000111888.3 |
Ccrl2
|
chemokine (C-C motif) receptor-like 2 |
| chr14_+_44340111 | 16.45 |
ENSMUST00000074839.7
|
Ear2
|
eosinophil-associated, ribonuclease A family, member 2 |
| chr16_+_49620883 | 16.40 |
ENSMUST00000229640.2
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr2_-_153083322 | 15.96 |
ENSMUST00000056924.14
|
Plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr5_-_24235295 | 15.75 |
ENSMUST00000101513.9
|
Fam126a
|
family with sequence similarity 126, member A |
| chr10_-_93727003 | 15.25 |
ENSMUST00000180840.8
|
Metap2
|
methionine aminopeptidase 2 |
| chr11_+_117673107 | 14.67 |
ENSMUST00000050874.14
ENSMUST00000106334.9 |
Tmc8
|
transmembrane channel-like gene family 8 |
| chr4_-_155012643 | 13.63 |
ENSMUST00000123514.8
|
Tnfrsf14
|
tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) |
| chr2_-_156848923 | 13.25 |
ENSMUST00000146413.8
ENSMUST00000103129.9 ENSMUST00000103130.8 |
Dsn1
|
DSN1 homolog, MIS12 kinetochore complex component |
| chr3_-_98800524 | 13.06 |
ENSMUST00000029464.9
|
Hao2
|
hydroxyacid oxidase 2 |
| chr5_-_24235646 | 13.02 |
ENSMUST00000197617.5
ENSMUST00000030849.13 |
Fam126a
|
family with sequence similarity 126, member A |
| chrX_+_55493325 | 12.80 |
ENSMUST00000079663.7
|
Gm2174
|
predicted gene 2174 |
| chr19_+_58748132 | 12.76 |
ENSMUST00000026081.5
|
Pnliprp2
|
pancreatic lipase-related protein 2 |
| chr17_+_34824827 | 12.53 |
ENSMUST00000037489.15
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) |
| chr16_-_19801781 | 12.37 |
ENSMUST00000058839.10
|
Klhl6
|
kelch-like 6 |
| chr4_-_41464816 | 12.14 |
ENSMUST00000108055.9
ENSMUST00000154535.8 ENSMUST00000030148.6 |
Kif24
|
kinesin family member 24 |
| chr7_+_97492124 | 12.10 |
ENSMUST00000033040.12
|
Pak1
|
p21 (RAC1) activated kinase 1 |
| chr6_-_25809209 | 12.08 |
ENSMUST00000115330.8
|
Pot1a
|
protection of telomeres 1A |
| chr6_+_30541581 | 11.90 |
ENSMUST00000096066.5
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr2_-_153067297 | 11.81 |
ENSMUST00000099194.4
|
Tspyl3
|
TSPY-like 3 |
| chr5_+_137628377 | 11.55 |
ENSMUST00000175968.8
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr11_+_58839716 | 11.18 |
ENSMUST00000078267.5
|
H2bu2
|
H2B.U histone 2 |
| chr15_+_57849269 | 10.64 |
ENSMUST00000050374.3
|
Fam83a
|
family with sequence similarity 83, member A |
| chr10_-_128462616 | 10.35 |
ENSMUST00000026420.7
|
Rps26
|
ribosomal protein S26 |
| chr11_+_117673198 | 10.08 |
ENSMUST00000117781.8
|
Tmc8
|
transmembrane channel-like gene family 8 |
| chr10_+_13376745 | 10.07 |
ENSMUST00000060212.13
ENSMUST00000121465.3 |
Fuca2
|
fucosidase, alpha-L- 2, plasma |
| chr16_+_58490625 | 9.98 |
ENSMUST00000060077.7
|
Cpox
|
coproporphyrinogen oxidase |
| chr2_-_102731691 | 9.58 |
ENSMUST00000111192.3
ENSMUST00000111190.9 ENSMUST00000111198.9 ENSMUST00000111191.9 ENSMUST00000060516.14 ENSMUST00000099673.9 ENSMUST00000005218.15 ENSMUST00000111194.8 |
Cd44
|
CD44 antigen |
| chr11_+_94827050 | 9.57 |
ENSMUST00000001547.8
|
Col1a1
|
collagen, type I, alpha 1 |
| chr14_+_22069877 | 9.55 |
ENSMUST00000161249.8
ENSMUST00000159777.8 ENSMUST00000162540.2 |
Lrmda
|
leucine rich melanocyte differentiation associated |
| chr10_-_80257681 | 9.46 |
ENSMUST00000156244.2
|
Tcf3
|
transcription factor 3 |
| chr8_-_78244412 | 9.10 |
ENSMUST00000210922.2
ENSMUST00000210519.2 |
Arhgap10
|
Rho GTPase activating protein 10 |
| chrX_-_137985960 | 8.86 |
ENSMUST00000033626.15
ENSMUST00000060824.4 |
Serpina7
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr4_-_56802266 | 8.80 |
ENSMUST00000030140.3
|
Elp1
|
elongator complex protein 1 |
| chr4_-_59438633 | 8.62 |
ENSMUST00000040166.14
ENSMUST00000107544.2 |
Susd1
|
sushi domain containing 1 |
| chr15_-_74635423 | 8.56 |
ENSMUST00000040404.8
|
Ly6d
|
lymphocyte antigen 6 complex, locus D |
| chr10_+_97400990 | 8.36 |
ENSMUST00000038160.6
|
Lum
|
lumican |
| chr9_-_110886576 | 8.24 |
ENSMUST00000199839.5
|
Ccrl2
|
chemokine (C-C motif) receptor-like 2 |
| chr6_-_25809188 | 8.20 |
ENSMUST00000115327.8
|
Pot1a
|
protection of telomeres 1A |
| chr13_-_95661726 | 8.18 |
ENSMUST00000022185.10
|
F2rl1
|
coagulation factor II (thrombin) receptor-like 1 |
| chr6_+_41279199 | 7.99 |
ENSMUST00000031913.5
|
Try4
|
trypsin 4 |
| chr5_-_137145030 | 7.95 |
ENSMUST00000054384.6
ENSMUST00000152207.2 |
Trim56
|
tripartite motif-containing 56 |
| chr11_-_117673008 | 7.90 |
ENSMUST00000152304.3
|
Tmc6
|
transmembrane channel-like gene family 6 |
| chrX_-_137985979 | 7.86 |
ENSMUST00000152457.2
|
Serpina7
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr6_-_34294377 | 7.67 |
ENSMUST00000154655.2
ENSMUST00000102980.11 |
Akr1b3
|
aldo-keto reductase family 1, member B3 (aldose reductase) |
| chr2_+_29951766 | 7.63 |
ENSMUST00000149578.8
|
Set
|
SET nuclear oncogene |
| chr9_+_66257747 | 7.48 |
ENSMUST00000042824.13
|
Herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr4_+_132262853 | 7.23 |
ENSMUST00000094657.10
ENSMUST00000105940.10 ENSMUST00000105939.10 ENSMUST00000150207.8 |
Dnajc8
|
DnaJ heat shock protein family (Hsp40) member C8 |
| chr6_+_67586695 | 7.18 |
ENSMUST00000103303.3
|
Igkv1-135
|
immunoglobulin kappa variable 1-135 |
| chr13_+_49835576 | 7.02 |
ENSMUST00000165316.8
ENSMUST00000047363.14 |
Iars
|
isoleucine-tRNA synthetase |
| chr2_+_126922156 | 6.99 |
ENSMUST00000142737.3
|
Blvra
|
biliverdin reductase A |
| chr1_-_23961379 | 6.85 |
ENSMUST00000027339.14
|
Smap1
|
small ArfGAP 1 |
| chr7_-_115445315 | 6.76 |
ENSMUST00000166207.3
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr7_+_16807965 | 6.62 |
ENSMUST00000071399.13
ENSMUST00000118367.3 |
Psg16
|
pregnancy specific glycoprotein 16 |
| chr3_+_89043879 | 6.53 |
ENSMUST00000107482.10
ENSMUST00000127058.2 |
Pklr
|
pyruvate kinase liver and red blood cell |
| chr3_+_89043440 | 6.50 |
ENSMUST00000047111.13
|
Pklr
|
pyruvate kinase liver and red blood cell |
| chr13_+_49835667 | 6.46 |
ENSMUST00000172254.3
|
Iars
|
isoleucine-tRNA synthetase |
| chr5_+_115983292 | 6.40 |
ENSMUST00000137952.8
ENSMUST00000148245.8 |
Cit
|
citron |
| chr4_+_56802338 | 6.30 |
ENSMUST00000045368.12
|
Abitram
|
actin binding transcription modulator |
| chr2_+_29951859 | 5.92 |
ENSMUST00000102866.10
|
Set
|
SET nuclear oncogene |
| chr11_-_78067446 | 5.85 |
ENSMUST00000148154.3
ENSMUST00000017549.13 |
Nek8
|
NIMA (never in mitosis gene a)-related expressed kinase 8 |
| chr7_+_142052569 | 5.83 |
ENSMUST00000078497.15
ENSMUST00000105953.10 ENSMUST00000179658.8 ENSMUST00000105954.10 ENSMUST00000105952.10 ENSMUST00000105955.8 ENSMUST00000074187.13 ENSMUST00000169299.9 ENSMUST00000105957.10 ENSMUST00000180152.8 ENSMUST00000105950.11 ENSMUST00000105958.10 ENSMUST00000105949.8 |
Tnnt3
|
troponin T3, skeletal, fast |
| chr11_+_117672902 | 5.60 |
ENSMUST00000127080.9
|
Tmc8
|
transmembrane channel-like gene family 8 |
| chr19_+_4608836 | 5.48 |
ENSMUST00000236457.2
|
Pcx
|
pyruvate carboxylase |
| chr14_+_22069709 | 5.47 |
ENSMUST00000075639.11
|
Lrmda
|
leucine rich melanocyte differentiation associated |
| chr9_+_99125420 | 5.31 |
ENSMUST00000185799.7
ENSMUST00000093795.10 ENSMUST00000190715.7 ENSMUST00000191335.7 ENSMUST00000190078.7 |
Cep70
|
centrosomal protein 70 |
| chr3_+_116653113 | 5.11 |
ENSMUST00000040260.11
|
Frrs1
|
ferric-chelate reductase 1 |
| chr6_+_117840031 | 5.00 |
ENSMUST00000172088.8
ENSMUST00000079405.15 |
Zfp239
|
zinc finger protein 239 |
| chr14_+_51366512 | 4.94 |
ENSMUST00000095923.4
|
Rnase6
|
ribonuclease, RNase A family, 6 |
| chr6_-_87567690 | 4.82 |
ENSMUST00000203636.2
ENSMUST00000050887.9 ENSMUST00000204682.3 |
Prokr1
|
prokineticin receptor 1 |
| chr10_-_117060377 | 4.81 |
ENSMUST00000020382.8
|
Yeats4
|
YEATS domain containing 4 |
| chr9_+_59198829 | 4.78 |
ENSMUST00000217570.2
ENSMUST00000026266.9 |
Adpgk
|
ADP-dependent glucokinase |
| chr9_-_106764627 | 4.42 |
ENSMUST00000055843.9
|
Rbm15b
|
RNA binding motif protein 15B |
| chr19_-_29344694 | 4.41 |
ENSMUST00000138051.2
|
Plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein |
| chr8_+_105066924 | 4.35 |
ENSMUST00000212081.2
|
Cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr17_-_26080429 | 4.34 |
ENSMUST00000079461.15
ENSMUST00000176923.9 |
Wdr90
|
WD repeat domain 90 |
| chr7_+_45683122 | 4.08 |
ENSMUST00000033121.7
|
Nomo1
|
nodal modulator 1 |
| chr19_+_29344846 | 4.06 |
ENSMUST00000016640.8
|
Cd274
|
CD274 antigen |
| chr1_-_171476495 | 3.94 |
ENSMUST00000194791.2
ENSMUST00000192024.6 |
Slamf7
|
SLAM family member 7 |
| chr1_-_171476559 | 3.84 |
ENSMUST00000111276.9
ENSMUST00000194531.6 |
Slamf7
|
SLAM family member 7 |
| chr4_+_41569775 | 3.81 |
ENSMUST00000102963.10
|
Dnai1
|
dynein axonemal intermediate chain 1 |
| chr8_+_108162985 | 3.81 |
ENSMUST00000166615.3
ENSMUST00000213097.2 ENSMUST00000212205.2 |
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr1_-_52271455 | 3.73 |
ENSMUST00000114512.8
|
Gls
|
glutaminase |
| chr3_+_88528743 | 3.66 |
ENSMUST00000170653.9
ENSMUST00000177303.8 |
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr18_-_36859732 | 3.57 |
ENSMUST00000061829.8
|
Cd14
|
CD14 antigen |
| chr6_-_38331482 | 3.46 |
ENSMUST00000031850.10
ENSMUST00000114898.3 |
Zc3hav1
|
zinc finger CCCH type, antiviral 1 |
| chr2_-_120561983 | 3.25 |
ENSMUST00000110701.8
ENSMUST00000110700.2 |
Cdan1
|
congenital dyserythropoietic anemia, type I (human) |
| chr7_+_4467730 | 3.19 |
ENSMUST00000086372.8
ENSMUST00000169820.8 ENSMUST00000163893.8 ENSMUST00000170635.2 |
Eps8l1
|
EPS8-like 1 |
| chr13_-_13568106 | 3.13 |
ENSMUST00000021738.10
ENSMUST00000220628.2 |
Gpr137b
|
G protein-coupled receptor 137B |
| chr3_-_83749036 | 3.08 |
ENSMUST00000029623.11
|
Tlr2
|
toll-like receptor 2 |
| chr14_-_44161016 | 2.99 |
ENSMUST00000159175.2
|
Ear10
|
eosinophil-associated, ribonuclease A family, member 10 |
| chr12_-_114477427 | 2.97 |
ENSMUST00000191803.2
|
Ighv1-5
|
immunoglobulin heavy variable V1-5 |
| chr2_+_181506130 | 2.95 |
ENSMUST00000039551.9
|
Polr3k
|
polymerase (RNA) III (DNA directed) polypeptide K |
| chr5_-_137202790 | 2.82 |
ENSMUST00000041226.11
|
Muc3
|
mucin 3, intestinal |
| chr2_-_152857239 | 2.82 |
ENSMUST00000028972.9
|
Pdrg1
|
p53 and DNA damage regulated 1 |
| chr6_+_125048230 | 2.75 |
ENSMUST00000140346.9
ENSMUST00000171989.3 |
Lpar5
|
lysophosphatidic acid receptor 5 |
| chr9_-_65734826 | 2.75 |
ENSMUST00000159109.2
|
Zfp609
|
zinc finger protein 609 |
| chr9_+_32547529 | 2.62 |
ENSMUST00000238819.2
|
Ets1
|
E26 avian leukemia oncogene 1, 5' domain |
| chr5_+_123482496 | 2.56 |
ENSMUST00000031391.9
ENSMUST00000117971.2 |
Bcl7a
|
B cell CLL/lymphoma 7A |
| chr13_-_74882328 | 2.55 |
ENSMUST00000223309.2
|
Cast
|
calpastatin |
| chr7_+_139900771 | 2.53 |
ENSMUST00000214594.2
|
Olfr525
|
olfactory receptor 525 |
| chr11_+_65698001 | 2.30 |
ENSMUST00000071465.9
ENSMUST00000018491.8 |
Zkscan6
|
zinc finger with KRAB and SCAN domains 6 |
| chr1_-_74076279 | 2.05 |
ENSMUST00000187281.7
|
Tns1
|
tensin 1 |
| chr2_+_143874979 | 2.04 |
ENSMUST00000037722.9
ENSMUST00000110032.2 |
Banf2
|
BANF family member 2 |
| chr1_+_171386752 | 2.01 |
ENSMUST00000004829.13
|
Cd244a
|
CD244 molecule A |
| chr15_+_12205095 | 2.01 |
ENSMUST00000038172.16
|
Mtmr12
|
myotubularin related protein 12 |
| chr1_-_131161312 | 1.93 |
ENSMUST00000212202.2
|
Rassf5
|
Ras association (RalGDS/AF-6) domain family member 5 |
| chr2_-_6726417 | 1.91 |
ENSMUST00000142941.8
ENSMUST00000150624.9 ENSMUST00000100429.11 ENSMUST00000182879.8 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chr17_-_13350642 | 1.90 |
ENSMUST00000232960.2
ENSMUST00000084966.6 ENSMUST00000233405.2 |
Unc93a
|
unc-93 homolog A |
| chr6_+_17306334 | 1.79 |
ENSMUST00000007799.13
ENSMUST00000115456.6 |
Cav1
|
caveolin 1, caveolae protein |
| chr2_-_166885414 | 1.69 |
ENSMUST00000067584.7
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr11_+_67090878 | 1.69 |
ENSMUST00000124516.8
ENSMUST00000018637.15 ENSMUST00000129018.8 |
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr2_-_87504008 | 1.66 |
ENSMUST00000213835.2
|
Olfr1135
|
olfactory receptor 1135 |
| chr14_+_64097739 | 1.59 |
ENSMUST00000022528.6
|
Pinx1
|
PIN2/TERF1 interacting, telomerase inhibitor 1 |
| chr2_-_20973692 | 1.57 |
ENSMUST00000114594.8
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr12_-_114528632 | 1.47 |
ENSMUST00000195337.2
ENSMUST00000103497.2 |
Ighv15-2
|
immunoglobulin heavy variable V15-2 |
| chr5_-_134205559 | 1.47 |
ENSMUST00000076228.3
|
Rcc1l
|
reculator of chromosome condensation 1 like |
| chr10_+_42378026 | 1.47 |
ENSMUST00000105500.8
ENSMUST00000019939.12 |
Snx3
|
sorting nexin 3 |
| chr12_-_108859123 | 1.41 |
ENSMUST00000161154.2
ENSMUST00000161410.8 |
Wars
|
tryptophanyl-tRNA synthetase |
| chr12_-_115884332 | 1.30 |
ENSMUST00000103548.3
|
Ighv1-81
|
immunoglobulin heavy variable 1-81 |
| chr9_-_105398346 | 1.26 |
ENSMUST00000176770.8
ENSMUST00000085133.13 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr10_+_80972089 | 1.25 |
ENSMUST00000048128.15
|
Zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr9_-_21042521 | 1.18 |
ENSMUST00000216874.2
ENSMUST00000214454.2 ENSMUST00000214864.2 |
Tyk2
|
tyrosine kinase 2 |
| chr10_+_80972368 | 1.14 |
ENSMUST00000119606.8
ENSMUST00000146895.2 ENSMUST00000121840.8 |
Zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr17_-_48739874 | 1.13 |
ENSMUST00000046549.5
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2 |
| chr16_+_17328924 | 1.12 |
ENSMUST00000232372.2
|
Lztr1
|
leucine-zipper-like transcriptional regulator, 1 |
| chr14_+_54713703 | 1.02 |
ENSMUST00000164697.8
|
Rem2
|
rad and gem related GTP binding protein 2 |
| chr18_+_89195089 | 1.01 |
ENSMUST00000236644.2
ENSMUST00000236828.2 |
Cd226
|
CD226 antigen |
| chr10_-_129962924 | 0.94 |
ENSMUST00000074161.2
|
Olfr824
|
olfactory receptor 824 |
| chr7_-_104983311 | 0.93 |
ENSMUST00000211006.3
ENSMUST00000216230.2 |
Olfr690
|
olfactory receptor 690 |
| chr7_+_97130135 | 0.79 |
ENSMUST00000026126.10
|
Ints4
|
integrator complex subunit 4 |
| chr9_+_39932760 | 0.70 |
ENSMUST00000215956.3
|
Olfr981
|
olfactory receptor 981 |
| chr1_-_173363523 | 0.68 |
ENSMUST00000139092.8
|
Ifi214
|
interferon activated gene 214 |
| chr8_+_105066980 | 0.64 |
ENSMUST00000211885.2
|
Cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr5_-_53864595 | 0.64 |
ENSMUST00000200691.4
|
Cckar
|
cholecystokinin A receptor |
| chr1_-_44140820 | 0.63 |
ENSMUST00000152239.8
|
Tex30
|
testis expressed 30 |
| chr6_+_48624158 | 0.60 |
ENSMUST00000203083.3
|
Gimap8
|
GTPase, IMAP family member 8 |
| chr4_-_118036827 | 0.56 |
ENSMUST00000097911.9
|
Kdm4a
|
lysine (K)-specific demethylase 4A |
| chr3_+_98129463 | 0.52 |
ENSMUST00000029469.5
|
Reg4
|
regenerating islet-derived family, member 4 |
| chr15_-_34356567 | 0.51 |
ENSMUST00000179647.2
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr6_-_68732577 | 0.47 |
ENSMUST00000103332.2
|
Igkv4-92
|
immunoglobulin kappa variable 4-92 |
| chr6_+_48624295 | 0.46 |
ENSMUST00000078223.6
ENSMUST00000203509.2 |
Gimap8
|
GTPase, IMAP family member 8 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 31.7 | 126.8 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 17.4 | 86.8 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 11.5 | 172.8 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 9.0 | 81.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 7.4 | 22.1 | GO:0034118 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 5.6 | 16.8 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 5.6 | 16.7 | GO:0033189 | response to vitamin A(GO:0033189) |
| 5.3 | 16.0 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 5.1 | 20.3 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 5.0 | 59.6 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 4.7 | 89.6 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 4.5 | 13.5 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 4.4 | 35.3 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 4.0 | 32.0 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 3.9 | 46.3 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 3.5 | 21.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 3.3 | 13.2 | GO:0051316 | attachment of spindle microtubules to kinetochore involved in meiotic chromosome segregation(GO:0051316) |
| 3.2 | 19.2 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 3.0 | 42.5 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 3.0 | 12.1 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 2.7 | 8.2 | GO:1902568 | positive regulation of eosinophil degranulation(GO:0043311) regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) positive regulation of renin secretion into blood stream(GO:1900135) positive regulation of eosinophil activation(GO:1902568) |
| 2.7 | 66.3 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 2.6 | 7.7 | GO:0090420 | hexitol metabolic process(GO:0006059) naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 2.4 | 9.6 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 2.4 | 94.2 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 2.2 | 39.6 | GO:0015816 | glycine transport(GO:0015816) |
| 2.2 | 55.9 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 2.0 | 10.0 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 2.0 | 19.6 | GO:0032264 | IMP salvage(GO:0032264) |
| 1.7 | 65.2 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 1.7 | 6.6 | GO:0071725 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 1.6 | 16.4 | GO:0008228 | opsonization(GO:0008228) |
| 1.6 | 17.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 1.3 | 13.0 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 1.2 | 9.5 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 1.2 | 19.7 | GO:0051601 | exocyst localization(GO:0051601) |
| 1.1 | 10.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 1.1 | 5.5 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 1.1 | 15.2 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 1.1 | 15.2 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 1.1 | 12.8 | GO:0019374 | galactolipid metabolic process(GO:0019374) |
| 1.0 | 30.3 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) |
| 1.0 | 8.4 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.9 | 37.8 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.9 | 7.0 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.9 | 19.9 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.9 | 13.6 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.8 | 20.9 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.8 | 59.0 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.8 | 6.8 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.7 | 28.8 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.7 | 12.5 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.7 | 36.1 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.7 | 22.5 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.6 | 11.6 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.6 | 4.4 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.6 | 3.7 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.6 | 4.1 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.5 | 1.6 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.5 | 5.0 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.5 | 22.5 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.5 | 3.7 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.4 | 5.8 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.4 | 7.5 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.4 | 13.1 | GO:0042537 | benzene-containing compound metabolic process(GO:0042537) |
| 0.3 | 19.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.3 | 15.0 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.3 | 2.6 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.3 | 1.3 | GO:0032468 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.3 | 1.6 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.3 | 12.4 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.3 | 3.5 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.3 | 3.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.2 | 23.5 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.2 | 6.4 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.2 | 6.8 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.2 | 1.0 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.2 | 18.7 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.2 | 4.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 12.1 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.2 | 1.0 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 13.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.2 | 0.6 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.1 | 2.8 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 2.6 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 21.1 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.1 | 3.2 | GO:0031497 | chromatin assembly(GO:0031497) |
| 0.1 | 8.7 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.1 | 2.7 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.1 | 3.0 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.1 | 6.0 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) |
| 0.1 | 1.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 9.5 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.1 | 5.2 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 3.8 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 0.2 | GO:0051977 | lysophospholipid transport(GO:0051977) |
| 0.1 | 4.8 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 2.9 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.6 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 5.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 2.0 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.8 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 2.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 1.1 | GO:0070232 | regulation of T cell apoptotic process(GO:0070232) |
| 0.0 | 15.7 | GO:0006935 | chemotaxis(GO:0006935) |
| 0.0 | 0.2 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.9 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.2 | 81.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 6.3 | 94.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 6.1 | 36.5 | GO:0032437 | cuticular plate(GO:0032437) |
| 5.7 | 80.2 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 4.4 | 13.2 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 3.2 | 9.6 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 2.0 | 20.3 | GO:0070187 | telosome(GO:0070187) |
| 1.9 | 40.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 1.9 | 20.9 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 1.6 | 19.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 1.5 | 7.7 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 1.5 | 5.8 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 1.3 | 26.3 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 1.1 | 28.8 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 1.0 | 3.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.9 | 174.4 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.8 | 13.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.8 | 8.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.8 | 19.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.8 | 32.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.6 | 9.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.6 | 12.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.6 | 12.8 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.5 | 20.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.4 | 3.6 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.4 | 19.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.3 | 2.8 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.2 | 111.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.2 | 24.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 10.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 4.8 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.2 | 2.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 12.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 3.0 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 13.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.5 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 10.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 36.3 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 7.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 6.4 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 23.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 18.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 3.7 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 159.6 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 11.6 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 52.7 | GO:0005768 | endosome(GO:0005768) |
| 0.1 | 1.7 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 9.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 81.1 | GO:0031410 | cytoplasmic vesicle(GO:0031410) |
| 0.0 | 0.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.6 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 3.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.5 | 86.8 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 10.7 | 32.0 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 9.1 | 36.5 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 6.8 | 20.3 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 6.4 | 44.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 5.5 | 22.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 5.0 | 19.9 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 4.6 | 141.2 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 4.5 | 18.0 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 4.5 | 13.5 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 4.4 | 35.3 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 4.1 | 20.3 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 3.8 | 45.6 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 3.4 | 10.1 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 3.3 | 13.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 2.9 | 172.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 2.9 | 28.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 2.8 | 39.6 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 2.7 | 87.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 2.6 | 13.0 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 2.4 | 26.3 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 2.1 | 19.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 2.0 | 8.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 2.0 | 19.6 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 1.9 | 19.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 1.5 | 21.0 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 1.5 | 16.4 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 1.4 | 19.6 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 1.4 | 7.0 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 1.4 | 5.5 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 1.3 | 24.7 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 1.2 | 9.5 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 1.1 | 89.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 1.0 | 3.1 | GO:0042497 | triacyl lipopeptide binding(GO:0042497) |
| 1.0 | 9.6 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 1.0 | 130.8 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.9 | 3.6 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.9 | 7.7 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.8 | 5.8 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) troponin I binding(GO:0031013) |
| 0.8 | 10.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.8 | 4.8 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.8 | 12.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.6 | 5.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.6 | 3.7 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.6 | 9.6 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.5 | 12.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.5 | 22.5 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.5 | 41.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.4 | 3.5 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.4 | 13.6 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.4 | 11.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.4 | 2.6 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.4 | 1.4 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.3 | 15.2 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.3 | 1.3 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.3 | 21.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.3 | 33.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.3 | 23.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.3 | 1.6 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.3 | 6.0 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 1.5 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.2 | 3.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.2 | 43.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 27.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 14.6 | GO:0042562 | hormone binding(GO:0042562) |
| 0.1 | 3.0 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 1.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 32.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 12.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 3.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 3.8 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 6.4 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 1.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 13.8 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.1 | 2.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 56.3 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.1 | 48.7 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.1 | 8.3 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.1 | 10.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 11.2 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) |
| 0.0 | 5.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 14.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 21.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.5 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 2.0 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 114.2 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 1.3 | 22.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 1.2 | 31.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 1.1 | 40.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.6 | 12.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.6 | 14.7 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.6 | 9.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.5 | 29.0 | PID EPO PATHWAY | EPO signaling pathway |
| 0.5 | 26.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.5 | 59.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.5 | 22.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.4 | 37.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.3 | 92.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.3 | 9.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.3 | 8.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.2 | 13.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 6.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 8.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 6.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 1.9 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 19.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 3.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 2.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 86.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 2.5 | 40.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 2.0 | 76.5 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 1.2 | 63.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 1.1 | 20.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 1.1 | 62.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 1.0 | 159.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.9 | 34.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.8 | 32.0 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.8 | 8.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.8 | 4.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.8 | 9.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.8 | 19.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.7 | 16.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.7 | 19.9 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.7 | 17.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.7 | 18.0 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.6 | 32.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.6 | 19.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.6 | 35.3 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.5 | 15.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.5 | 14.9 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.4 | 13.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.3 | 3.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.3 | 9.0 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.3 | 3.6 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.2 | 3.7 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 12.3 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.2 | 10.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 9.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 13.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 3.0 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 8.9 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.1 | 1.2 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 13.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 5.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 3.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |