Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Tbx19
Z-value: 2.04
Transcription factors associated with Tbx19
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx19
|
ENSMUSG00000026572.12 | Tbx19 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx19 | mm39_v1_chr1_-_164988342_164988350 | -0.43 | 9.3e-03 | Click! |
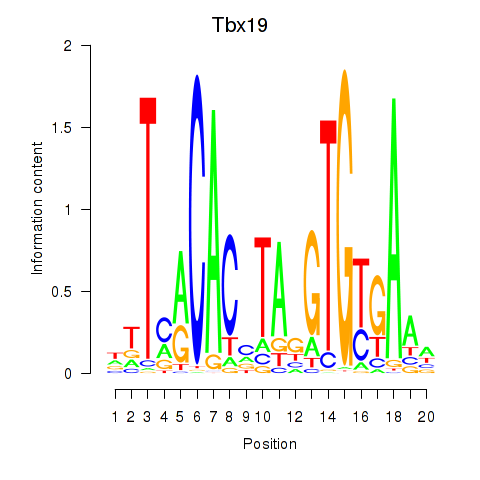
Activity profile of Tbx19 motif
Sorted Z-values of Tbx19 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx19
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_138121245 | 17.94 |
ENSMUST00000161312.8
ENSMUST00000013458.9 |
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr4_-_60455331 | 13.65 |
ENSMUST00000135953.2
|
Mup1
|
major urinary protein 1 |
| chr19_-_40175709 | 13.24 |
ENSMUST00000051846.13
|
Cyp2c70
|
cytochrome P450, family 2, subfamily c, polypeptide 70 |
| chr13_+_4484305 | 12.09 |
ENSMUST00000021630.15
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chr2_+_24944407 | 8.29 |
ENSMUST00000102931.11
ENSMUST00000074422.14 ENSMUST00000132172.8 ENSMUST00000114388.8 |
Nsmf
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr9_-_118986123 | 8.25 |
ENSMUST00000010795.5
|
Acaa1b
|
acetyl-Coenzyme A acyltransferase 1B |
| chr3_+_94600863 | 8.14 |
ENSMUST00000090848.10
ENSMUST00000173981.8 ENSMUST00000173849.8 ENSMUST00000174223.2 |
Selenbp2
|
selenium binding protein 2 |
| chr10_-_24803336 | 7.41 |
ENSMUST00000020161.10
|
Arg1
|
arginase, liver |
| chr19_+_12610668 | 7.41 |
ENSMUST00000044976.12
|
Glyat
|
glycine-N-acyltransferase |
| chr3_+_59989282 | 6.67 |
ENSMUST00000029326.6
|
Sucnr1
|
succinate receptor 1 |
| chr10_-_93375832 | 6.10 |
ENSMUST00000016034.3
|
Amdhd1
|
amidohydrolase domain containing 1 |
| chr5_+_45650821 | 5.72 |
ENSMUST00000198534.2
|
Lap3
|
leucine aminopeptidase 3 |
| chr9_-_103107495 | 5.53 |
ENSMUST00000035158.16
|
Trf
|
transferrin |
| chr11_+_101358990 | 5.48 |
ENSMUST00000001347.7
|
Rnd2
|
Rho family GTPase 2 |
| chr18_-_56695333 | 4.97 |
ENSMUST00000066208.13
ENSMUST00000172734.8 |
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr19_-_58443830 | 4.68 |
ENSMUST00000026076.14
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr1_+_185064339 | 4.67 |
ENSMUST00000027916.13
ENSMUST00000210277.2 ENSMUST00000151769.2 ENSMUST00000110965.2 |
Bpnt1
|
3'(2'), 5'-bisphosphate nucleotidase 1 |
| chr11_-_59937302 | 4.54 |
ENSMUST00000000310.14
ENSMUST00000102693.9 ENSMUST00000148512.2 |
Pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr8_-_110305672 | 4.50 |
ENSMUST00000074898.8
|
Hp
|
haptoglobin |
| chrM_+_10167 | 4.35 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr19_-_58443593 | 4.33 |
ENSMUST00000135730.2
ENSMUST00000152507.8 |
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr17_+_79934096 | 4.23 |
ENSMUST00000224618.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr6_+_129157576 | 3.97 |
ENSMUST00000032260.6
|
Clec2d
|
C-type lectin domain family 2, member d |
| chr14_-_123150497 | 3.92 |
ENSMUST00000162164.2
ENSMUST00000110679.9 ENSMUST00000161322.3 ENSMUST00000038075.12 |
Ggact
|
gamma-glutamylamine cyclotransferase |
| chr9_-_103107460 | 3.92 |
ENSMUST00000165296.8
ENSMUST00000112645.8 |
Trf
|
transferrin |
| chr6_-_119444157 | 3.84 |
ENSMUST00000118120.8
|
Wnt5b
|
wingless-type MMTV integration site family, member 5B |
| chrM_+_9870 | 3.84 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr8_-_41586713 | 3.67 |
ENSMUST00000155055.2
ENSMUST00000059115.13 ENSMUST00000145860.2 |
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr12_-_81014849 | 3.65 |
ENSMUST00000095572.5
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr10_-_81262948 | 3.62 |
ENSMUST00000078185.14
ENSMUST00000020461.15 ENSMUST00000105321.10 |
Nfic
|
nuclear factor I/C |
| chr17_+_37253802 | 3.60 |
ENSMUST00000040498.12
|
Rnf39
|
ring finger protein 39 |
| chr6_+_41279199 | 3.49 |
ENSMUST00000031913.5
|
Try4
|
trypsin 4 |
| chr4_+_155646807 | 3.41 |
ENSMUST00000030939.14
|
Nadk
|
NAD kinase |
| chr12_-_81014755 | 3.35 |
ENSMUST00000218342.2
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr1_-_72251466 | 3.23 |
ENSMUST00000048860.9
|
Mreg
|
melanoregulin |
| chr19_+_41017714 | 3.04 |
ENSMUST00000051806.12
ENSMUST00000112200.3 |
Dntt
|
deoxynucleotidyltransferase, terminal |
| chr19_+_8814782 | 3.04 |
ENSMUST00000171649.8
|
Bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr14_-_55822696 | 2.94 |
ENSMUST00000022828.9
|
Emc9
|
ER membrane protein complex subunit 9 |
| chr6_+_90442269 | 2.88 |
ENSMUST00000113530.4
|
Klf15
|
Kruppel-like factor 15 |
| chr11_-_101062111 | 2.88 |
ENSMUST00000164474.8
ENSMUST00000043397.14 |
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr2_-_112198366 | 2.79 |
ENSMUST00000028551.4
|
Emc4
|
ER membrane protein complex subunit 4 |
| chr14_-_34032311 | 2.66 |
ENSMUST00000111917.3
ENSMUST00000228704.2 |
Shld2
|
shieldin complex subunit 2 |
| chr15_+_31602252 | 2.52 |
ENSMUST00000042702.7
ENSMUST00000161061.3 |
Atpsckmt
|
ATP synthase C subunit lysine N-methyltransferase |
| chrX_+_72830607 | 2.44 |
ENSMUST00000166518.8
|
Ssr4
|
signal sequence receptor, delta |
| chr17_+_56935118 | 2.42 |
ENSMUST00000112979.4
|
Catsperd
|
cation channel sperm associated auxiliary subunit delta |
| chr17_+_7246289 | 2.40 |
ENSMUST00000179728.2
|
Rnaset2b
|
ribonuclease T2B |
| chr16_+_48662894 | 2.39 |
ENSMUST00000238847.2
ENSMUST00000023329.7 |
Retnla
|
resistin like alpha |
| chr10_+_88295515 | 2.19 |
ENSMUST00000125612.2
|
Sycp3
|
synaptonemal complex protein 3 |
| chr13_+_58954447 | 2.16 |
ENSMUST00000224259.2
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr17_-_8366536 | 2.10 |
ENSMUST00000231927.2
|
Rnaset2a
|
ribonuclease T2A |
| chr19_+_56276343 | 2.10 |
ENSMUST00000095948.11
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr5_-_115622356 | 2.07 |
ENSMUST00000112067.8
|
Sirt4
|
sirtuin 4 |
| chr5_+_34683141 | 2.05 |
ENSMUST00000125817.8
ENSMUST00000067638.14 |
Sh3bp2
|
SH3-domain binding protein 2 |
| chr4_-_12087911 | 2.00 |
ENSMUST00000050686.10
|
Tmem67
|
transmembrane protein 67 |
| chr19_+_56276375 | 1.98 |
ENSMUST00000166049.8
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr7_-_140535828 | 1.94 |
ENSMUST00000211129.2
|
Gm45717
|
predicted gene 45717 |
| chr5_-_38649291 | 1.90 |
ENSMUST00000129099.8
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr5_+_100054110 | 1.84 |
ENSMUST00000198837.3
|
Vamp9
|
vesicle-associated membrane protein 9 |
| chr19_+_12824046 | 1.76 |
ENSMUST00000189517.2
|
Gm5244
|
predicted pseudogene 5244 |
| chr19_+_34268053 | 1.73 |
ENSMUST00000025691.13
|
Fas
|
Fas (TNF receptor superfamily member 6) |
| chr5_+_52521133 | 1.73 |
ENSMUST00000101208.6
|
Sod3
|
superoxide dismutase 3, extracellular |
| chr17_+_7246365 | 1.70 |
ENSMUST00000232245.2
|
Rnaset2b
|
ribonuclease T2B |
| chr14_+_66378382 | 1.70 |
ENSMUST00000022620.11
|
Chrna2
|
cholinergic receptor, nicotinic, alpha polypeptide 2 (neuronal) |
| chr9_+_108447077 | 1.67 |
ENSMUST00000019183.14
|
Dalrd3
|
DALR anticodon binding domain containing 3 |
| chr14_+_30673334 | 1.56 |
ENSMUST00000226551.2
ENSMUST00000228328.2 |
Nek4
|
NIMA (never in mitosis gene a)-related expressed kinase 4 |
| chr7_-_19415301 | 1.56 |
ENSMUST00000150569.9
ENSMUST00000127648.4 ENSMUST00000003071.10 |
Gm44805
Apoc4
|
predicted gene 44805 apolipoprotein C-IV |
| chr14_+_14091030 | 1.52 |
ENSMUST00000224529.2
|
Oit1
|
oncoprotein induced transcript 1 |
| chr7_+_28441400 | 1.51 |
ENSMUST00000094632.6
|
Sars2
|
seryl-aminoacyl-tRNA synthetase 2 |
| chr4_-_107540726 | 1.48 |
ENSMUST00000131776.8
|
Dmrtb1
|
DMRT-like family B with proline-rich C-terminal, 1 |
| chr1_-_59134042 | 1.47 |
ENSMUST00000238601.2
ENSMUST00000238949.2 ENSMUST00000097080.4 |
C2cd6
|
C2 calcium dependent domain containing 6 |
| chr2_-_147888816 | 1.47 |
ENSMUST00000172928.2
ENSMUST00000047315.10 |
Foxa2
|
forkhead box A2 |
| chr17_+_25992761 | 1.46 |
ENSMUST00000237541.2
|
Ciao3
|
cytosolic iron-sulfur assembly component 3 |
| chr14_+_14090981 | 1.45 |
ENSMUST00000022269.7
|
Oit1
|
oncoprotein induced transcript 1 |
| chr11_+_72192455 | 1.43 |
ENSMUST00000151440.8
ENSMUST00000146233.8 ENSMUST00000140842.9 |
Xaf1
|
XIAP associated factor 1 |
| chr1_+_33758937 | 1.41 |
ENSMUST00000088287.10
|
Rab23
|
RAB23, member RAS oncogene family |
| chr17_+_25992742 | 1.39 |
ENSMUST00000134108.8
ENSMUST00000002350.11 |
Ciao3
|
cytosolic iron-sulfur assembly component 3 |
| chr19_+_34268071 | 1.35 |
ENSMUST00000112472.4
ENSMUST00000235232.2 |
Fas
|
Fas (TNF receptor superfamily member 6) |
| chr13_+_3588063 | 1.35 |
ENSMUST00000223396.2
ENSMUST00000059515.8 ENSMUST00000222365.2 |
Gdi2
|
guanosine diphosphate (GDP) dissociation inhibitor 2 |
| chr4_-_21767116 | 1.35 |
ENSMUST00000029915.6
|
Tstd3
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 3 |
| chr2_+_36575800 | 1.34 |
ENSMUST00000213258.2
|
Olfr346
|
olfactory receptor 346 |
| chr11_-_113540867 | 1.32 |
ENSMUST00000136392.8
ENSMUST00000125890.8 ENSMUST00000146031.8 |
Slc39a11
|
solute carrier family 39 (metal ion transporter), member 11 |
| chr4_+_122730027 | 1.25 |
ENSMUST00000030412.11
ENSMUST00000121870.8 ENSMUST00000097902.5 |
Ppt1
|
palmitoyl-protein thioesterase 1 |
| chr8_+_105951777 | 1.16 |
ENSMUST00000034361.10
|
D230025D16Rik
|
RIKEN cDNA D230025D16 gene |
| chr4_+_155789246 | 1.15 |
ENSMUST00000030905.9
|
Ssu72
|
Ssu72 RNA polymerase II CTD phosphatase homolog (yeast) |
| chr10_-_85847697 | 1.13 |
ENSMUST00000105304.2
ENSMUST00000061699.12 |
Bpifc
|
BPI fold containing family C |
| chr13_+_58954374 | 1.12 |
ENSMUST00000225488.2
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr7_-_140535899 | 1.09 |
ENSMUST00000081649.10
|
Ifitm2
|
interferon induced transmembrane protein 2 |
| chr2_+_153716958 | 1.09 |
ENSMUST00000028983.3
|
Bpifb2
|
BPI fold containing family B, member 2 |
| chr13_+_55875158 | 1.08 |
ENSMUST00000021958.6
ENSMUST00000124968.8 |
Pcbd2
|
pterin 4 alpha carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 2 |
| chr17_+_37253916 | 1.08 |
ENSMUST00000173072.2
|
Rnf39
|
ring finger protein 39 |
| chrX_+_139565657 | 1.08 |
ENSMUST00000112990.8
ENSMUST00000112988.8 |
Mid2
|
midline 2 |
| chr10_+_88295431 | 1.03 |
ENSMUST00000020252.10
|
Sycp3
|
synaptonemal complex protein 3 |
| chr16_-_44566700 | 1.03 |
ENSMUST00000023348.11
ENSMUST00000162512.8 |
Gtpbp8
|
GTP-binding protein 8 (putative) |
| chr1_+_183170293 | 1.01 |
ENSMUST00000192076.3
|
Taf1a
|
TATA-box binding protein associated factor, RNA polymerase I, A |
| chr16_-_44566641 | 0.98 |
ENSMUST00000161436.2
|
Gtpbp8
|
GTP-binding protein 8 (putative) |
| chr2_-_155534235 | 0.98 |
ENSMUST00000103140.5
|
Trpc4ap
|
transient receptor potential cation channel, subfamily C, member 4 associated protein |
| chr19_-_10897993 | 0.95 |
ENSMUST00000025641.2
|
Zp1
|
zona pellucida glycoprotein 1 |
| chr6_+_86172196 | 0.93 |
ENSMUST00000032066.13
|
Tgfa
|
transforming growth factor alpha |
| chr15_+_39997761 | 0.92 |
ENSMUST00000228780.2
|
9330182O14Rik
|
RIKEN cDNA 9330182O14 gene |
| chr7_+_143383814 | 0.90 |
ENSMUST00000141916.8
ENSMUST00000144034.8 ENSMUST00000143338.2 ENSMUST00000207143.2 ENSMUST00000125564.2 |
Dhcr7
|
7-dehydrocholesterol reductase |
| chr9_-_16289527 | 0.90 |
ENSMUST00000082170.6
|
Fat3
|
FAT atypical cadherin 3 |
| chr7_+_48608800 | 0.87 |
ENSMUST00000183659.8
|
Nav2
|
neuron navigator 2 |
| chr1_+_161322219 | 0.86 |
ENSMUST00000086084.2
|
Tnfsf18
|
tumor necrosis factor (ligand) superfamily, member 18 |
| chr11_+_73262072 | 0.85 |
ENSMUST00000078952.9
ENSMUST00000120401.9 ENSMUST00000170592.4 |
Olfr376
|
olfactory receptor 376 |
| chr17_+_46957151 | 0.85 |
ENSMUST00000002844.14
ENSMUST00000113429.8 ENSMUST00000113430.2 |
Mrpl2
|
mitochondrial ribosomal protein L2 |
| chr13_+_93440265 | 0.79 |
ENSMUST00000109494.8
|
Homer1
|
homer scaffolding protein 1 |
| chr9_+_48896765 | 0.75 |
ENSMUST00000047349.8
|
Usp28
|
ubiquitin specific peptidase 28 |
| chrX_-_55867668 | 0.73 |
ENSMUST00000135542.2
ENSMUST00000114766.8 |
Map7d3
|
MAP7 domain containing 3 |
| chr10_+_81919146 | 0.73 |
ENSMUST00000220287.2
|
BC024063
|
cDNA sequence BC024063 |
| chr8_+_117231712 | 0.71 |
ENSMUST00000213007.2
ENSMUST00000078170.7 |
Dynlrb2
|
dynein light chain roadblock-type 2 |
| chr11_+_69217078 | 0.70 |
ENSMUST00000018614.3
|
Kcnab3
|
potassium voltage-gated channel, shaker-related subfamily, beta member 3 |
| chrX_-_100865583 | 0.68 |
ENSMUST00000239206.2
|
Gm3858
|
predicted gene 3858 |
| chr7_-_103113358 | 0.68 |
ENSMUST00000214347.2
|
Olfr607
|
olfactory receptor 607 |
| chr5_+_98328723 | 0.66 |
ENSMUST00000112959.4
|
Prdm8
|
PR domain containing 8 |
| chr19_-_3464447 | 0.63 |
ENSMUST00000025842.8
ENSMUST00000237521.2 |
Gal
|
galanin and GMAP prepropeptide |
| chr7_-_24145107 | 0.62 |
ENSMUST00000205776.2
|
Irgc1
|
immunity-related GTPase family, cinema 1 |
| chr6_+_86986818 | 0.61 |
ENSMUST00000032060.15
ENSMUST00000117583.8 ENSMUST00000144776.7 |
Nfu1
|
NFU1 iron-sulfur cluster scaffold |
| chr7_-_126613707 | 0.61 |
ENSMUST00000165096.9
|
Mvp
|
major vault protein |
| chr1_-_39844467 | 0.60 |
ENSMUST00000171319.4
|
Gm3646
|
predicted gene 3646 |
| chr7_-_29605949 | 0.58 |
ENSMUST00000159920.2
ENSMUST00000162592.8 |
Zfp27
|
zinc finger protein 27 |
| chrX_+_32070863 | 0.57 |
ENSMUST00000238237.2
|
Btbd35f1
|
BTB domain containing 35, family member 1 |
| chr4_-_84464521 | 0.57 |
ENSMUST00000177040.2
|
Bnc2
|
basonuclin 2 |
| chrX_-_33014777 | 0.56 |
ENSMUST00000186329.2
|
Btbd35f15
|
BTB domain containing 35, family member 15 |
| chrX_-_33139812 | 0.56 |
ENSMUST00000105117.3
|
Btbd35f14
|
BTB domain containing 35, family member 14 |
| chrX_+_31839202 | 0.56 |
ENSMUST00000179991.3
|
Btbd35f2
|
BTB domain containing 35, family member 2 |
| chr12_+_103564479 | 0.55 |
ENSMUST00000190151.2
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chrX_-_31034822 | 0.55 |
ENSMUST00000238426.2
|
Btbd35f19
|
BTB domain containing 35, family member 19 |
| chr9_+_107440445 | 0.54 |
ENSMUST00000010198.5
|
Tusc2
|
tumor suppressor 2, mitochondrial calcium regulator |
| chr10_+_3784877 | 0.54 |
ENSMUST00000239159.2
|
Plekhg1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chrX_+_32411401 | 0.53 |
ENSMUST00000178747.3
|
Btbd35f5
|
BTB domain containing 35, family member 5 |
| chrX_+_32750842 | 0.53 |
ENSMUST00000178827.3
|
Btbd35f12
|
BTB domain containing 35, family member 12 |
| chr17_-_33435325 | 0.52 |
ENSMUST00000112162.4
|
Olfr1564
|
olfactory receptor 1564 |
| chr10_+_81963832 | 0.51 |
ENSMUST00000201286.4
|
AU041133
|
expressed sequence AU041133 |
| chrX_-_33394003 | 0.51 |
ENSMUST00000179466.2
|
Btbd35f6
|
BTB domain containing 35, family member 6 |
| chrX_+_30768610 | 0.51 |
ENSMUST00000179532.2
|
Btbd35f29
|
BTB domain containing 35, family member 29 |
| chrX_+_33094635 | 0.50 |
ENSMUST00000177912.2
|
Btbd35f13
|
BTB domain containing 35, family member 13 |
| chr8_+_93628015 | 0.50 |
ENSMUST00000104947.5
|
Capns2
|
calpain, small subunit 2 |
| chr7_-_5548308 | 0.50 |
ENSMUST00000236262.2
|
Vmn1r60
|
vomeronasal 1 receptor 60 |
| chr5_-_143123955 | 0.47 |
ENSMUST00000218872.3
|
Olfr718-ps1
|
olfactory receptor 718, pseudogene 1 |
| chrX_-_4194587 | 0.46 |
ENSMUST00000179325.2
|
Btbd35f20
|
BTB domain containing 35, family member 20 |
| chr7_+_12661337 | 0.45 |
ENSMUST00000045870.5
|
Rnf225
|
ring finger protein 225 |
| chr1_+_127796508 | 0.45 |
ENSMUST00000037649.6
ENSMUST00000212506.2 |
Rab3gap1
|
RAB3 GTPase activating protein subunit 1 |
| chr14_-_66071337 | 0.44 |
ENSMUST00000225853.2
|
Esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr17_-_57348647 | 0.44 |
ENSMUST00000169012.2
ENSMUST00000058661.14 |
Slc25a41
|
solute carrier family 25, member 41 |
| chr9_-_25063068 | 0.44 |
ENSMUST00000008573.9
|
Herpud2
|
HERPUD family member 2 |
| chr2_-_174909556 | 0.42 |
ENSMUST00000072895.10
ENSMUST00000109066.2 |
Gm14393
|
predicted gene 14393 |
| chr14_+_65903840 | 0.42 |
ENSMUST00000022610.15
|
Scara5
|
scavenger receptor class A, member 5 |
| chr6_+_56933451 | 0.41 |
ENSMUST00000096612.4
|
Vmn1r4
|
vomeronasal 1 receptor 4 |
| chr14_+_20753074 | 0.40 |
ENSMUST00000071215.5
ENSMUST00000224633.2 |
Chchd1
|
coiled-coil-helix-coiled-coil-helix domain containing 1 |
| chrX_-_33580888 | 0.39 |
ENSMUST00000238632.2
|
Btbd35f9
|
BTB domain containing 35, family member 9 |
| chrX_+_132809189 | 0.39 |
ENSMUST00000113304.2
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chrX_-_3150852 | 0.39 |
ENSMUST00000178080.3
|
Btbd35f24
|
BTB domain containing 35, family member 24 |
| chrX_-_3398715 | 0.39 |
ENSMUST00000105020.2
|
Btbd35f11
|
BTB domain containing 35, family member 11 |
| chrX_+_30987337 | 0.39 |
ENSMUST00000238765.2
|
Btbd35f26
|
BTB domain containing 35, family member 26 |
| chr3_-_88280047 | 0.38 |
ENSMUST00000107543.8
ENSMUST00000107542.2 |
Bglap3
|
bone gamma-carboxyglutamate protein 3 |
| chr2_-_177269992 | 0.37 |
ENSMUST00000108945.8
ENSMUST00000108943.2 |
Gm14406
|
predicted gene 14406 |
| chr2_+_174852034 | 0.37 |
ENSMUST00000109069.8
ENSMUST00000109070.3 |
Gm14444
|
predicted gene 14444 |
| chrX_+_3605258 | 0.35 |
ENSMUST00000105019.3
|
Btbd35f18
|
BTB domain containing 35, family member 18 |
| chrX_-_3657910 | 0.35 |
ENSMUST00000178621.2
|
Btbd35f10
|
BTB domain containing 35, family member 10 |
| chr7_+_17546558 | 0.35 |
ENSMUST00000023953.5
|
Ceacam14
|
carcinoembryonic antigen-related cell adhesion molecule 14 |
| chrX_+_31608531 | 0.35 |
ENSMUST00000238528.2
|
Btbd35f22
|
BTB domain containing 35, family member 22 |
| chrX_+_4273978 | 0.33 |
ENSMUST00000105014.2
|
Btbd35f17
|
BTB domain containing 35, family member 17 |
| chr6_+_128911336 | 0.32 |
ENSMUST00000000254.14
|
Clec2g
|
C-type lectin domain family 2, member g |
| chr16_-_45664664 | 0.32 |
ENSMUST00000036355.13
|
Phldb2
|
pleckstrin homology like domain, family B, member 2 |
| chr7_-_133966588 | 0.32 |
ENSMUST00000172947.8
|
D7Ertd443e
|
DNA segment, Chr 7, ERATO Doi 443, expressed |
| chrX_+_4101601 | 0.31 |
ENSMUST00000105015.3
|
Btbd35f28
|
BTB domain containing 35, family member 28 |
| chrX_-_3862027 | 0.31 |
ENSMUST00000185755.2
|
Btbd35f16
|
BTB domain containing 35, family member 16 |
| chr5_-_129781323 | 0.31 |
ENSMUST00000042266.13
|
Septin14
|
septin 14 |
| chr7_-_102540089 | 0.30 |
ENSMUST00000217024.2
|
Olfr569
|
olfactory receptor 569 |
| chr4_+_62443606 | 0.29 |
ENSMUST00000062145.2
|
4933430I17Rik
|
RIKEN cDNA 4933430I17 gene |
| chr1_-_161615927 | 0.29 |
ENSMUST00000193648.2
|
Fasl
|
Fas ligand (TNF superfamily, member 6) |
| chr12_-_113771372 | 0.29 |
ENSMUST00000103456.4
|
Ighv2-7
|
immunoglobulin heavy variable 2-7 |
| chr10_-_25412010 | 0.29 |
ENSMUST00000179685.3
|
Smlr1
|
small leucine-rich protein 1 |
| chrX_+_11178173 | 0.28 |
ENSMUST00000178979.2
|
H2al1e
|
H2A histone family member L1E |
| chr1_-_69726384 | 0.27 |
ENSMUST00000187184.7
|
Ikzf2
|
IKAROS family zinc finger 2 |
| chr1_+_34537450 | 0.26 |
ENSMUST00000027299.10
|
Prss39
|
protease, serine 39 |
| chr14_-_63781381 | 0.24 |
ENSMUST00000058679.7
|
Mtmr9
|
myotubularin related protein 9 |
| chr14_+_53886861 | 0.24 |
ENSMUST00000103660.4
|
Trav15-2-dv6-2
|
T cell receptor alpha variable 15-2-DV6-2 |
| chr2_-_113588983 | 0.23 |
ENSMUST00000099575.4
|
Grem1
|
gremlin 1, DAN family BMP antagonist |
| chrX_+_132809166 | 0.23 |
ENSMUST00000033606.15
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chr7_+_17706049 | 0.22 |
ENSMUST00000094799.3
|
Ceacam11
|
carcinoembryonic antigen-related cell adhesion molecule 11 |
| chr5_-_46013838 | 0.21 |
ENSMUST00000087164.10
ENSMUST00000121573.8 |
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr4_+_143341573 | 0.20 |
ENSMUST00000105773.2
|
Pramel21
|
PRAME like 21 |
| chr1_-_119765068 | 0.20 |
ENSMUST00000163435.8
|
Ptpn4
|
protein tyrosine phosphatase, non-receptor type 4 |
| chr7_-_7124478 | 0.19 |
ENSMUST00000056246.8
|
Zfp954
|
zinc finger protein 954 |
| chr1_-_161616031 | 0.17 |
ENSMUST00000000834.4
|
Fasl
|
Fas ligand (TNF superfamily, member 6) |
| chr2_-_88937158 | 0.16 |
ENSMUST00000099806.3
ENSMUST00000213288.2 |
Olfr1220
|
olfactory receptor 1220 |
| chr7_+_39167154 | 0.14 |
ENSMUST00000108015.4
|
Zfp619
|
zinc finger protein 619 |
| chr6_+_17749169 | 0.13 |
ENSMUST00000053148.14
ENSMUST00000115417.4 |
St7
|
suppression of tumorigenicity 7 |
| chr2_-_177016100 | 0.13 |
ENSMUST00000108959.3
|
Gm14412
|
predicted gene 14412 |
| chrX_+_11184495 | 0.13 |
ENSMUST00000179859.2
|
H2al1g
|
H2A histone family member L1G |
| chr11_-_115212851 | 0.11 |
ENSMUST00000103037.5
|
Ush1g
|
USH1 protein network component sans |
| chrX_+_32236876 | 0.10 |
ENSMUST00000105017.5
|
Btbd35f21
|
BTB domain containing 35, family member 21 |
| chr6_+_24857967 | 0.10 |
ENSMUST00000200968.4
|
Hyal5
|
hyaluronoglucosaminidase 5 |
| chr4_+_43384320 | 0.09 |
ENSMUST00000136360.2
|
Rusc2
|
RUN and SH3 domain containing 2 |
| chr5_+_138185747 | 0.09 |
ENSMUST00000110934.9
|
Cnpy4
|
canopy FGF signaling regulator 4 |
| chr7_-_89914610 | 0.09 |
ENSMUST00000107221.9
ENSMUST00000107220.8 ENSMUST00000040413.2 |
Ccdc83
|
coiled-coil domain containing 83 |
| chr9_-_67336085 | 0.08 |
ENSMUST00000213584.2
|
Tln2
|
talin 2 |
| chr13_-_22368139 | 0.08 |
ENSMUST00000237107.2
|
Vmn1r191
|
vomeronasal 1 receptor 191 |
| chr14_+_65903878 | 0.06 |
ENSMUST00000069226.7
|
Scara5
|
scavenger receptor class A, member 5 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 17.9 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 4.0 | 12.1 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 1.5 | 4.5 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 1.5 | 7.4 | GO:0043091 | regulation of amino acid import(GO:0010958) L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 1.4 | 8.3 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 1.2 | 6.1 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 1.2 | 9.4 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 1.1 | 13.6 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 1.1 | 3.2 | GO:0051878 | lateral element assembly(GO:0051878) |
| 1.0 | 3.1 | GO:0031104 | dendrite regeneration(GO:0031104) |
| 0.7 | 5.0 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.5 | 3.3 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.5 | 1.6 | GO:1900062 | regulation of replicative cell aging(GO:1900062) |
| 0.5 | 1.5 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.5 | 4.0 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.5 | 1.5 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.5 | 2.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.4 | 2.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.4 | 3.4 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.4 | 11.7 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.3 | 5.5 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.3 | 0.9 | GO:0016131 | brassinosteroid metabolic process(GO:0016131) brassinosteroid biosynthetic process(GO:0016132) |
| 0.3 | 0.9 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.2 | 4.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 1.3 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 1.6 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.2 | 6.0 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.2 | 0.5 | GO:0052572 | interleukin-15 production(GO:0032618) response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.2 | 0.6 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.2 | 2.0 | GO:0035845 | negative regulation of centrosome duplication(GO:0010826) photoreceptor cell outer segment organization(GO:0035845) negative regulation of centrosome cycle(GO:0046606) |
| 0.2 | 3.0 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.6 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.1 | 0.9 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 4.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.5 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 3.9 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.1 | 3.2 | GO:0042640 | anagen(GO:0042640) |
| 0.1 | 1.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.1 | 1.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 2.9 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 | 1.4 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 3.6 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 0.4 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 2.2 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 1.9 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 3.8 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 3.0 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 0.2 | GO:1900158 | negative regulation of monocyte chemotaxis(GO:0090027) negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.1 | 2.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 1.0 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.1 | 1.7 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.6 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 3.8 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.6 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.9 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 1.7 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.8 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 9.0 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation(GO:0050731) |
| 0.0 | 0.7 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.0 | 3.6 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 6.3 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 1.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.6 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 1.9 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.9 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.6 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 3.5 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 1.4 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.0 | 1.2 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 8.3 | GO:0006631 | fatty acid metabolic process(GO:0006631) |
| 0.0 | 0.5 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 1.2 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.8 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.3 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.5 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.9 | 9.4 | GO:0097433 | dense body(GO:0097433) |
| 0.6 | 3.2 | GO:0000802 | transverse filament(GO:0000802) |
| 0.4 | 5.7 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.3 | 2.4 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.3 | 3.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.2 | 2.4 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.2 | 1.0 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.2 | 8.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 2.0 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 6.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 8.2 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 1.0 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 1.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 1.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 1.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 1.6 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 9.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 3.0 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 2.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.9 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 9.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 4.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 7.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 8.3 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 5.7 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 7.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.8 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 1.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 1.2 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 2.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 17.9 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 4.5 | 13.6 | GO:0005009 | insulin-activated receptor activity(GO:0005009) |
| 4.0 | 12.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 1.9 | 7.4 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 1.3 | 9.4 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 1.2 | 5.0 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 1.2 | 4.7 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.9 | 7.0 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.8 | 3.1 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.7 | 6.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.7 | 7.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.7 | 3.9 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.6 | 4.5 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.6 | 8.3 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.5 | 3.3 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.5 | 1.5 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.5 | 8.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.4 | 4.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.4 | 1.1 | GO:0008124 | 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.3 | 1.4 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.3 | 11.7 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.3 | 0.9 | GO:0047598 | sterol delta7 reductase activity(GO:0009918) 7-dehydrocholesterol reductase activity(GO:0047598) |
| 0.3 | 1.7 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.3 | 8.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 1.3 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.2 | 4.3 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 1.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 3.4 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 1.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 1.9 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 2.1 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.6 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 7.3 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 0.5 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 0.1 | 0.4 | GO:0008147 | structural constituent of bone(GO:0008147) |
| 0.1 | 3.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) acetylcholine binding(GO:0042166) |
| 0.1 | 0.7 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 2.0 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 3.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 8.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.8 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 9.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 6.2 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.4 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.4 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 7.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 2.0 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 1.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.5 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 1.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 2.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.7 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 1.2 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 12.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 3.0 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 4.7 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 5.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 7.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 3.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.4 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 2.1 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 3.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 4.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 17.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.6 | 7.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 4.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 3.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 9.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 4.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.7 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 7.4 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 18.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 1.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 3.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 2.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 3.3 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |