Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
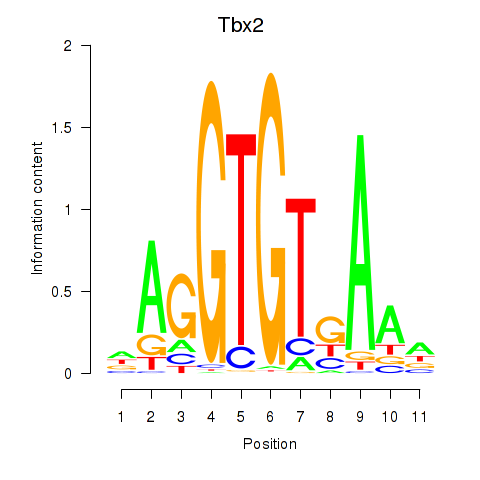
Results for Tbx2
Z-value: 2.38
Transcription factors associated with Tbx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx2
|
ENSMUSG00000000093.7 | Tbx2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx2 | mm39_v1_chr11_+_85723377_85723377 | -0.61 | 6.8e-05 | Click! |
Activity profile of Tbx2 motif
Sorted Z-values of Tbx2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_130754413 | 25.88 |
ENSMUST00000027675.14
ENSMUST00000133792.8 |
Pigr
|
polymeric immunoglobulin receptor |
| chr17_-_34218301 | 16.35 |
ENSMUST00000235463.2
|
H2-K1
|
histocompatibility 2, K1, K region |
| chr19_+_20470056 | 15.75 |
ENSMUST00000225337.3
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr15_+_6474808 | 14.97 |
ENSMUST00000022749.17
ENSMUST00000239466.2 |
C9
|
complement component 9 |
| chr6_-_141801897 | 14.03 |
ENSMUST00000165990.8
|
Slco1a4
|
solute carrier organic anion transporter family, member 1a4 |
| chr19_+_20470114 | 12.39 |
ENSMUST00000225313.2
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr6_-_141801918 | 11.07 |
ENSMUST00000163678.2
|
Slco1a4
|
solute carrier organic anion transporter family, member 1a4 |
| chr9_-_43151179 | 8.43 |
ENSMUST00000034512.7
|
Oaf
|
out at first homolog |
| chr16_+_22926162 | 7.38 |
ENSMUST00000023599.13
ENSMUST00000168891.8 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr19_+_37686240 | 7.35 |
ENSMUST00000025946.7
|
Cyp26a1
|
cytochrome P450, family 26, subfamily a, polypeptide 1 |
| chr1_-_51955126 | 7.21 |
ENSMUST00000046390.14
|
Myo1b
|
myosin IB |
| chr14_-_30665232 | 6.78 |
ENSMUST00000006704.17
ENSMUST00000163118.2 |
Itih1
|
inter-alpha trypsin inhibitor, heavy chain 1 |
| chr3_-_107851021 | 5.88 |
ENSMUST00000106684.8
ENSMUST00000106685.9 |
Gstm6
|
glutathione S-transferase, mu 6 |
| chr6_-_21851827 | 5.82 |
ENSMUST00000202353.2
ENSMUST00000134635.2 ENSMUST00000123116.8 ENSMUST00000120965.8 ENSMUST00000143531.2 |
Tspan12
|
tetraspanin 12 |
| chr6_+_116241146 | 5.45 |
ENSMUST00000112900.9
ENSMUST00000036503.14 ENSMUST00000223495.2 |
Zfand4
|
zinc finger, AN1-type domain 4 |
| chr5_-_151113619 | 4.48 |
ENSMUST00000062015.15
ENSMUST00000110483.9 |
Stard13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr3_-_115508680 | 4.45 |
ENSMUST00000055676.4
|
S1pr1
|
sphingosine-1-phosphate receptor 1 |
| chr1_+_88066086 | 4.43 |
ENSMUST00000014263.6
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr8_-_106660470 | 4.10 |
ENSMUST00000034368.8
|
Ctrl
|
chymotrypsin-like |
| chr4_+_84802592 | 4.04 |
ENSMUST00000102819.10
|
Cntln
|
centlein, centrosomal protein |
| chr5_-_5564873 | 3.92 |
ENSMUST00000060947.14
|
Cldn12
|
claudin 12 |
| chr5_-_5564730 | 3.89 |
ENSMUST00000115445.8
ENSMUST00000179804.8 ENSMUST00000125110.2 ENSMUST00000115446.8 |
Cldn12
|
claudin 12 |
| chr7_-_45136235 | 3.68 |
ENSMUST00000210701.2
|
Gm45808
|
predicted gene 45808 |
| chr4_+_84802513 | 3.66 |
ENSMUST00000047023.13
|
Cntln
|
centlein, centrosomal protein |
| chr4_-_49549489 | 3.66 |
ENSMUST00000029987.10
|
Aldob
|
aldolase B, fructose-bisphosphate |
| chr15_-_81244940 | 3.59 |
ENSMUST00000023040.9
|
Slc25a17
|
solute carrier family 25 (mitochondrial carrier, peroxisomal membrane protein), member 17 |
| chr1_+_131898325 | 3.59 |
ENSMUST00000027695.8
|
Slc45a3
|
solute carrier family 45, member 3 |
| chrX_+_100427331 | 3.48 |
ENSMUST00000119190.2
|
Gjb1
|
gap junction protein, beta 1 |
| chr19_-_42190589 | 3.14 |
ENSMUST00000018966.8
|
Sfrp5
|
secreted frizzled-related sequence protein 5 |
| chr4_+_41465134 | 3.09 |
ENSMUST00000030154.7
|
Nudt2
|
nudix (nucleoside diphosphate linked moiety X)-type motif 2 |
| chr6_-_94677118 | 2.92 |
ENSMUST00000101126.3
ENSMUST00000032105.11 |
Lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr3_+_59939175 | 2.76 |
ENSMUST00000029325.5
|
Aadac
|
arylacetamide deacetylase |
| chr3_+_90421742 | 2.62 |
ENSMUST00000048138.8
|
S100a13
|
S100 calcium binding protein A13 |
| chr5_+_120651158 | 2.62 |
ENSMUST00000111889.2
|
Slc8b1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr14_-_55880708 | 2.62 |
ENSMUST00000120041.8
ENSMUST00000121937.8 ENSMUST00000133707.2 ENSMUST00000002391.15 ENSMUST00000121791.8 |
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr14_+_11307729 | 2.49 |
ENSMUST00000160956.2
ENSMUST00000160340.8 ENSMUST00000162278.8 |
Fhit
|
fragile histidine triad gene |
| chr12_-_75782502 | 2.44 |
ENSMUST00000021450.6
|
Sgpp1
|
sphingosine-1-phosphate phosphatase 1 |
| chr15_-_96917804 | 2.42 |
ENSMUST00000231039.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr3_+_52175757 | 2.41 |
ENSMUST00000053764.7
|
Foxo1
|
forkhead box O1 |
| chr18_+_36414122 | 2.41 |
ENSMUST00000051301.6
|
Pura
|
purine rich element binding protein A |
| chr10_+_106306122 | 2.35 |
ENSMUST00000029404.17
ENSMUST00000217854.2 |
Ppfia2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr2_+_160722562 | 2.31 |
ENSMUST00000109456.9
|
Lpin3
|
lipin 3 |
| chr3_+_85878376 | 2.31 |
ENSMUST00000238443.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr7_+_19197192 | 2.26 |
ENSMUST00000137613.9
|
Exoc3l2
|
exocyst complex component 3-like 2 |
| chr13_+_81034214 | 2.22 |
ENSMUST00000161441.2
|
Arrdc3
|
arrestin domain containing 3 |
| chr9_-_20657643 | 2.17 |
ENSMUST00000215999.2
|
Olfm2
|
olfactomedin 2 |
| chr13_+_112600604 | 2.13 |
ENSMUST00000183663.8
ENSMUST00000184311.8 ENSMUST00000183886.8 |
Il6st
|
interleukin 6 signal transducer |
| chr9_+_48406706 | 2.11 |
ENSMUST00000048824.9
|
Gm5617
|
predicted gene 5617 |
| chr4_+_116078787 | 2.07 |
ENSMUST00000147292.8
|
Pik3r3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr4_+_116078874 | 2.02 |
ENSMUST00000106490.3
|
Pik3r3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr9_-_63509699 | 2.00 |
ENSMUST00000171243.2
ENSMUST00000163982.8 ENSMUST00000163624.8 |
Iqch
|
IQ motif containing H |
| chr7_+_27307422 | 1.99 |
ENSMUST00000142365.8
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr2_+_52747855 | 1.98 |
ENSMUST00000155586.9
ENSMUST00000090952.11 ENSMUST00000127122.9 ENSMUST00000049483.14 ENSMUST00000050719.13 |
Fmnl2
|
formin-like 2 |
| chr6_+_22288220 | 1.96 |
ENSMUST00000128245.8
ENSMUST00000031681.10 ENSMUST00000148639.2 |
Wnt16
|
wingless-type MMTV integration site family, member 16 |
| chr4_-_140393185 | 1.96 |
ENSMUST00000069623.12
|
Arhgef10l
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr7_-_28001624 | 1.92 |
ENSMUST00000108315.4
|
Dll3
|
delta like canonical Notch ligand 3 |
| chr16_-_38533597 | 1.88 |
ENSMUST00000023487.5
|
Arhgap31
|
Rho GTPase activating protein 31 |
| chr1_+_132243849 | 1.86 |
ENSMUST00000072177.14
ENSMUST00000082125.6 |
Nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chrX_-_138683102 | 1.84 |
ENSMUST00000101217.4
|
Ripply1
|
ripply transcriptional repressor 1 |
| chr7_+_105053775 | 1.81 |
ENSMUST00000033187.6
ENSMUST00000210344.2 |
Cnga4
|
cyclic nucleotide gated channel alpha 4 |
| chr15_-_95426108 | 1.80 |
ENSMUST00000075275.3
|
Nell2
|
NEL-like 2 |
| chr7_+_27770655 | 1.79 |
ENSMUST00000138392.8
ENSMUST00000076648.8 |
Fcgbp
|
Fc fragment of IgG binding protein |
| chr7_+_4240697 | 1.73 |
ENSMUST00000117550.2
|
Lilra5
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 |
| chr1_-_105284383 | 1.70 |
ENSMUST00000058688.7
|
Rnf152
|
ring finger protein 152 |
| chr17_+_46991972 | 1.63 |
ENSMUST00000002845.8
|
Mea1
|
male enhanced antigen 1 |
| chr9_-_63509747 | 1.53 |
ENSMUST00000080527.12
ENSMUST00000042322.11 |
Iqch
|
IQ motif containing H |
| chr9_-_48406564 | 1.49 |
ENSMUST00000213276.2
ENSMUST00000170000.4 |
Rbm7
|
RNA binding motif protein 7 |
| chr1_+_87254729 | 1.48 |
ENSMUST00000172794.8
ENSMUST00000164992.9 ENSMUST00000173173.8 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr7_-_30262512 | 1.46 |
ENSMUST00000207747.2
ENSMUST00000207797.2 |
Psenen
|
presenilin enhancer gamma secretase subunit |
| chr19_+_8817883 | 1.44 |
ENSMUST00000086058.13
|
Bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr1_-_80439165 | 1.35 |
ENSMUST00000211023.2
|
Gm45261
|
predicted gene 45261 |
| chr4_+_116078830 | 1.30 |
ENSMUST00000030464.14
|
Pik3r3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr6_+_50087826 | 1.29 |
ENSMUST00000167628.2
|
Mpp6
|
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) |
| chr1_-_105284407 | 1.29 |
ENSMUST00000172299.2
|
Rnf152
|
ring finger protein 152 |
| chr7_+_27307173 | 1.29 |
ENSMUST00000136962.8
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr5_+_96357337 | 1.28 |
ENSMUST00000117766.8
|
Mrpl1
|
mitochondrial ribosomal protein L1 |
| chr11_-_23720953 | 1.28 |
ENSMUST00000102864.5
|
Rel
|
reticuloendotheliosis oncogene |
| chr7_+_133239414 | 1.27 |
ENSMUST00000128901.9
|
Edrf1
|
erythroid differentiation regulatory factor 1 |
| chr1_-_87017633 | 1.23 |
ENSMUST00000027455.13
ENSMUST00000188310.2 |
Alppl2
|
alkaline phosphatase, placental-like 2 |
| chr10_+_97528915 | 1.23 |
ENSMUST00000060703.6
|
Ccer1
|
coiled-coil glutamate-rich protein 1 |
| chr1_+_163942833 | 1.22 |
ENSMUST00000162746.2
|
Selp
|
selectin, platelet |
| chr15_-_76004395 | 1.22 |
ENSMUST00000239552.1
|
EPPK1
|
epiplakin 1 |
| chr1_+_92838479 | 1.22 |
ENSMUST00000027487.15
|
Rnpepl1
|
arginyl aminopeptidase (aminopeptidase B)-like 1 |
| chr5_-_3852857 | 1.21 |
ENSMUST00000043551.11
|
Ankib1
|
ankyrin repeat and IBR domain containing 1 |
| chr1_+_10108433 | 1.21 |
ENSMUST00000071087.12
ENSMUST00000117415.8 |
Cspp1
|
centrosome and spindle pole associated protein 1 |
| chr11_+_98441998 | 1.20 |
ENSMUST00000107513.3
|
Zpbp2
|
zona pellucida binding protein 2 |
| chr1_-_171910324 | 1.14 |
ENSMUST00000003550.11
|
Ncstn
|
nicastrin |
| chr1_+_171910073 | 1.11 |
ENSMUST00000135192.8
|
Copa
|
coatomer protein complex subunit alpha |
| chr5_-_87634665 | 1.10 |
ENSMUST00000201519.2
|
Gm43638
|
predicted gene 43638 |
| chr9_-_22042930 | 1.08 |
ENSMUST00000213815.2
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr15_-_67048595 | 1.06 |
ENSMUST00000229213.2
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr10_+_98750268 | 1.05 |
ENSMUST00000219557.2
|
Atp2b1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr7_+_79460475 | 1.02 |
ENSMUST00000107394.3
|
Mesp2
|
mesoderm posterior 2 |
| chr4_-_119031050 | 1.00 |
ENSMUST00000052715.10
ENSMUST00000179290.8 ENSMUST00000154226.2 |
Zfp691
|
zinc finger protein 691 |
| chr1_-_156936197 | 0.94 |
ENSMUST00000187546.7
ENSMUST00000118207.8 ENSMUST00000027884.13 ENSMUST00000121911.8 |
Tex35
|
testis expressed 35 |
| chr7_-_30826184 | 0.93 |
ENSMUST00000211945.2
|
Scn1b
|
sodium channel, voltage-gated, type I, beta |
| chr11_-_23845207 | 0.93 |
ENSMUST00000102863.3
ENSMUST00000020513.10 |
Papolg
|
poly(A) polymerase gamma |
| chr14_+_52131067 | 0.93 |
ENSMUST00000047726.12
|
Slc39a2
|
solute carrier family 39 (zinc transporter), member 2 |
| chr2_-_103627937 | 0.91 |
ENSMUST00000028607.13
|
Caprin1
|
cell cycle associated protein 1 |
| chr4_+_156214969 | 0.86 |
ENSMUST00000209248.2
|
Rnf223
|
ring finger 223 |
| chr9_-_22043083 | 0.86 |
ENSMUST00000069330.14
ENSMUST00000217643.2 |
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr17_+_71859026 | 0.85 |
ENSMUST00000124001.8
ENSMUST00000167641.8 ENSMUST00000064420.12 |
Spdya
|
speedy/RINGO cell cycle regulator family, member A |
| chr13_+_33268095 | 0.85 |
ENSMUST00000016951.8
|
Serpinb1b
|
serine (or cysteine) peptidase inhibitor, clade B, member 1b |
| chr1_+_152830720 | 0.84 |
ENSMUST00000043313.15
ENSMUST00000186621.2 |
Nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr1_+_171910343 | 0.83 |
ENSMUST00000027833.12
|
Copa
|
coatomer protein complex subunit alpha |
| chr4_+_94502719 | 0.83 |
ENSMUST00000107104.3
ENSMUST00000030311.11 |
Ift74
|
intraflagellar transport 74 |
| chr2_+_172821620 | 0.82 |
ENSMUST00000109125.8
ENSMUST00000109126.5 ENSMUST00000050442.15 |
Spo11
|
SPO11 initiator of meiotic double stranded breaks |
| chr14_+_50683002 | 0.82 |
ENSMUST00000214792.2
|
Olfr740
|
olfactory receptor 740 |
| chr1_+_87254719 | 0.81 |
ENSMUST00000027475.15
|
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr19_+_28941292 | 0.78 |
ENSMUST00000045674.4
|
Plpp6
|
phospholipid phosphatase 6 |
| chr6_+_42263609 | 0.77 |
ENSMUST00000238845.2
ENSMUST00000031894.13 |
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chr11_+_98441923 | 0.76 |
ENSMUST00000081033.13
ENSMUST00000107511.8 ENSMUST00000107509.8 ENSMUST00000017339.12 |
Zpbp2
|
zona pellucida binding protein 2 |
| chr6_+_42263644 | 0.74 |
ENSMUST00000163936.8
|
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chr10_+_90412114 | 0.70 |
ENSMUST00000182427.8
ENSMUST00000182053.8 ENSMUST00000182113.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr19_+_21249636 | 0.69 |
ENSMUST00000237651.2
|
Zfand5
|
zinc finger, AN1-type domain 5 |
| chr1_-_172156828 | 0.68 |
ENSMUST00000194204.2
|
Kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr3_-_145355725 | 0.66 |
ENSMUST00000029846.5
|
Ccn1
|
cellular communication network factor 1 |
| chr3_+_96011810 | 0.64 |
ENSMUST00000132980.8
ENSMUST00000138206.8 ENSMUST00000090785.9 ENSMUST00000035519.12 |
Otud7b
|
OTU domain containing 7B |
| chr15_-_89294434 | 0.61 |
ENSMUST00000109314.9
|
Syce3
|
synaptonemal complex central element protein 3 |
| chr15_-_67048673 | 0.61 |
ENSMUST00000229028.2
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr1_-_172156884 | 0.61 |
ENSMUST00000062387.8
|
Kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr14_+_53574579 | 0.59 |
ENSMUST00000179580.3
|
Trav13n-3
|
T cell receptor alpha variable 13N-3 |
| chr16_-_36648586 | 0.58 |
ENSMUST00000023537.6
ENSMUST00000114829.9 |
Eaf2
|
ELL associated factor 2 |
| chr5_+_52521133 | 0.58 |
ENSMUST00000101208.6
|
Sod3
|
superoxide dismutase 3, extracellular |
| chr6_-_28261881 | 0.57 |
ENSMUST00000115320.8
ENSMUST00000123098.8 ENSMUST00000115321.9 ENSMUST00000155494.2 |
Zfp800
|
zinc finger protein 800 |
| chr1_+_132119169 | 0.55 |
ENSMUST00000188169.7
ENSMUST00000112357.9 ENSMUST00000188175.2 |
Lemd1
Gm29695
|
LEM domain containing 1 predicted gene, 29695 |
| chr1_-_161615927 | 0.54 |
ENSMUST00000193648.2
|
Fasl
|
Fas ligand (TNF superfamily, member 6) |
| chr12_-_87742525 | 0.52 |
ENSMUST00000164517.3
|
Eif1ad19
|
eukaryotic translation initiation factor 1A domain containing 19 |
| chr7_+_45135784 | 0.52 |
ENSMUST00000107758.10
|
Tulp2
|
tubby-like protein 2 |
| chr19_-_28941264 | 0.52 |
ENSMUST00000161813.2
|
Spata6l
|
spermatogenesis associated 6 like |
| chr17_-_16051295 | 0.52 |
ENSMUST00000231985.2
|
Rgmb
|
repulsive guidance molecule family member B |
| chr1_+_93301596 | 0.50 |
ENSMUST00000058682.11
ENSMUST00000186641.7 |
Ano7
|
anoctamin 7 |
| chr1_+_131526977 | 0.50 |
ENSMUST00000027690.7
|
Avpr1b
|
arginine vasopressin receptor 1B |
| chr2_-_164670452 | 0.48 |
ENSMUST00000017911.4
|
Spata25
|
spermatogenesis associated 25 |
| chr19_+_4053290 | 0.48 |
ENSMUST00000025806.5
|
Doc2g
|
double C2, gamma |
| chr4_+_125384481 | 0.47 |
ENSMUST00000030676.8
|
Grik3
|
glutamate receptor, ionotropic, kainate 3 |
| chr10_-_62723103 | 0.47 |
ENSMUST00000218438.2
|
Tet1
|
tet methylcytosine dioxygenase 1 |
| chrX_+_159865500 | 0.45 |
ENSMUST00000238603.2
|
Scml2
|
Scm polycomb group protein like 2 |
| chr11_-_83959175 | 0.45 |
ENSMUST00000100705.11
|
Dusp14
|
dual specificity phosphatase 14 |
| chr4_+_53440389 | 0.44 |
ENSMUST00000107646.9
ENSMUST00000102911.10 |
Slc44a1
|
solute carrier family 44, member 1 |
| chr4_+_148085179 | 0.44 |
ENSMUST00000103230.5
|
Nppa
|
natriuretic peptide type A |
| chr2_+_130038463 | 0.44 |
ENSMUST00000166774.3
|
Tmc2
|
transmembrane channel-like gene family 2 |
| chrX_+_99773784 | 0.43 |
ENSMUST00000113744.2
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr9_+_63509925 | 0.43 |
ENSMUST00000041551.9
|
Aagab
|
alpha- and gamma-adaptin binding protein |
| chr15_+_79113341 | 0.41 |
ENSMUST00000163571.8
|
Pick1
|
protein interacting with C kinase 1 |
| chr3_-_108062172 | 0.39 |
ENSMUST00000062028.8
|
Gpr61
|
G protein-coupled receptor 61 |
| chr4_+_127062924 | 0.39 |
ENSMUST00000046659.14
|
Dlgap3
|
DLG associated protein 3 |
| chrX_-_133375735 | 0.34 |
ENSMUST00000113223.3
|
Taf7l
|
TATA-box binding protein associated factor 7 like |
| chr1_-_190711151 | 0.33 |
ENSMUST00000047409.9
|
Vash2
|
vasohibin 2 |
| chr2_-_60503998 | 0.32 |
ENSMUST00000059888.15
ENSMUST00000154764.2 |
Itgb6
|
integrin beta 6 |
| chr4_-_58785722 | 0.31 |
ENSMUST00000059608.5
|
Olfr267
|
olfactory receptor 267 |
| chr11_-_87783073 | 0.31 |
ENSMUST00000213672.2
ENSMUST00000213928.2 |
Olfr462
|
olfactory receptor 462 |
| chr9_-_85631361 | 0.29 |
ENSMUST00000039213.15
|
Ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chrX_-_47123719 | 0.28 |
ENSMUST00000039026.8
|
Apln
|
apelin |
| chr14_+_68321302 | 0.28 |
ENSMUST00000022639.8
|
Nefl
|
neurofilament, light polypeptide |
| chr7_-_141783708 | 0.26 |
ENSMUST00000097942.3
|
Krtap5-5
|
keratin associated protein 5-5 |
| chr7_-_121306476 | 0.26 |
ENSMUST00000046929.7
|
Usp31
|
ubiquitin specific peptidase 31 |
| chrX_+_151789457 | 0.24 |
ENSMUST00000095755.4
|
Usp51
|
ubiquitin specific protease 51 |
| chr2_-_91013362 | 0.22 |
ENSMUST00000066420.12
|
Madd
|
MAP-kinase activating death domain |
| chr2_+_68691902 | 0.21 |
ENSMUST00000176018.2
|
Cers6
|
ceramide synthase 6 |
| chr7_-_108529375 | 0.19 |
ENSMUST00000055745.5
|
Nlrp10
|
NLR family, pyrin domain containing 10 |
| chr6_-_24956296 | 0.18 |
ENSMUST00000127247.4
|
Tmem229a
|
transmembrane protein 229A |
| chr9_-_40915895 | 0.17 |
ENSMUST00000180384.3
|
Crtam
|
cytotoxic and regulatory T cell molecule |
| chrX_+_99773523 | 0.16 |
ENSMUST00000019503.14
|
Gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr1_-_161616031 | 0.15 |
ENSMUST00000000834.4
|
Fasl
|
Fas ligand (TNF superfamily, member 6) |
| chr18_+_65158873 | 0.15 |
ENSMUST00000226058.2
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr6_-_141954471 | 0.14 |
ENSMUST00000111832.3
|
Gm6614
|
predicted gene 6614 |
| chr8_-_26505605 | 0.13 |
ENSMUST00000016138.11
|
Fnta
|
farnesyltransferase, CAAX box, alpha |
| chr1_+_171156568 | 0.12 |
ENSMUST00000111300.8
|
Dedd
|
death effector domain-containing |
| chr9_+_110948492 | 0.11 |
ENSMUST00000217341.3
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr12_-_52074846 | 0.11 |
ENSMUST00000040161.5
|
Gpr33
|
G protein-coupled receptor 33 |
| chr16_-_32688640 | 0.10 |
ENSMUST00000089684.10
ENSMUST00000040986.15 ENSMUST00000115105.9 |
Rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr10_-_23968192 | 0.10 |
ENSMUST00000092654.4
|
Taar8b
|
trace amine-associated receptor 8B |
| chr7_-_79443536 | 0.09 |
ENSMUST00000032760.6
|
Mesp1
|
mesoderm posterior 1 |
| chr15_-_99864936 | 0.08 |
ENSMUST00000100209.6
|
Fam186a
|
family with sequence similarity 186, member A |
| chr2_-_120946877 | 0.08 |
ENSMUST00000110675.3
|
Tgm7
|
transglutaminase 7 |
| chr2_+_151923449 | 0.07 |
ENSMUST00000064061.4
|
Scrt2
|
scratch family zinc finger 2 |
| chrX_+_41591410 | 0.07 |
ENSMUST00000005839.11
|
Sh2d1a
|
SH2 domain containing 1A |
| chr11_-_69791774 | 0.07 |
ENSMUST00000102580.10
|
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr10_+_26105605 | 0.05 |
ENSMUST00000218301.2
ENSMUST00000164660.8 ENSMUST00000060716.6 |
Samd3
|
sterile alpha motif domain containing 3 |
| chr16_-_88609108 | 0.05 |
ENSMUST00000232664.2
|
Gm20741
|
predicted gene, 20741 |
| chr4_+_119397710 | 0.04 |
ENSMUST00000160219.2
|
Foxj3
|
forkhead box J3 |
| chr7_+_26456567 | 0.03 |
ENSMUST00000077855.8
|
Cyp2b19
|
cytochrome P450, family 2, subfamily b, polypeptide 19 |
| chr9_-_40915858 | 0.02 |
ENSMUST00000188848.8
ENSMUST00000034519.13 |
Crtam
|
cytotoxic and regulatory T cell molecule |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.6 | 25.9 | GO:0002414 | immune response in mucosal-associated lymphoid tissue(GO:0002386) immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 3.3 | 16.4 | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 3.1 | 28.1 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 1.5 | 4.5 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 1.1 | 7.4 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 1.0 | 3.1 | GO:2000040 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 1.0 | 2.9 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.9 | 3.6 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.9 | 4.5 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.9 | 15.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.7 | 2.2 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.7 | 2.1 | GO:0070104 | negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.6 | 7.1 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.6 | 3.7 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.5 | 25.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.5 | 3.3 | GO:0071484 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.4 | 7.7 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.4 | 2.4 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.4 | 2.0 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.4 | 1.9 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.4 | 4.4 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.3 | 0.9 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.3 | 2.6 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.3 | 2.5 | GO:0015961 | diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.3 | 0.8 | GO:1990918 | meiotic DNA double-strand break formation(GO:0042138) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.2 | 1.9 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 | 1.3 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.2 | 2.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 2.8 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.2 | 0.7 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.2 | 0.5 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.2 | 2.4 | GO:0071455 | cellular response to hyperoxia(GO:0071455) |
| 0.2 | 1.5 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 6.8 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 2.6 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.1 | 1.3 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 | 0.8 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 5.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 1.1 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 1.0 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 7.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 2.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 2.2 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 0.5 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.1 | 2.6 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 0.8 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 1.8 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 0.4 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.1 | 0.6 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 5.5 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 0.3 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.1 | 0.3 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.1 | 2.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 2.0 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 2.1 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.1 | 1.2 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.1 | 2.0 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.4 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 0.4 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.5 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 | 0.4 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.7 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 1.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.2 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.1 | 7.2 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.1 | 0.6 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 2.3 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.6 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 3.0 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.4 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 1.8 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 2.3 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.3 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.5 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.5 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 1.2 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.7 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 1.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 1.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.6 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.2 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.0 | 0.1 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.0 | 0.9 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.9 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.9 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.7 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 0.6 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 1.2 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 15.0 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 1.2 | 16.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.7 | 3.7 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.7 | 2.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.6 | 25.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.3 | 1.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 1.8 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.2 | 7.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 2.6 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 0.6 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.2 | 1.3 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.2 | 3.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 5.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 2.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 3.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.9 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 2.6 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.4 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.1 | 2.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 3.6 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 1.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 2.0 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 2.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 7.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.3 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.1 | 0.6 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 2.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 4.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 5.4 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.8 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 25.7 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 0.4 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 2.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 3.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 28.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 2.2 | 25.9 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 1.2 | 7.4 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 1.1 | 25.1 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 1.1 | 16.4 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.9 | 3.6 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.7 | 3.6 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.7 | 2.1 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.7 | 2.6 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.6 | 2.5 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.6 | 2.4 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.6 | 2.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.6 | 2.2 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.5 | 5.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.4 | 9.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 0.9 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.3 | 1.2 | GO:0042806 | fucose binding(GO:0042806) |
| 0.3 | 1.8 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.3 | 7.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.3 | 4.5 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.3 | 2.6 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.2 | 3.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 0.8 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.2 | 1.7 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.2 | 0.9 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 1.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.2 | 3.1 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.2 | 1.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 3.7 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.1 | 1.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.8 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 2.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 4.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 4.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.9 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 5.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 2.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.6 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 7.4 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 0.5 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.4 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 1.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.5 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 2.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.6 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.5 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 0.6 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 5.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 1.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 7.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.4 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 2.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.3 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 1.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 1.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.7 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.5 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 1.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.7 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 2.7 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.8 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 5.9 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 2.0 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 3.0 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 1.5 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 2.2 | GO:0030674 | protein binding, bridging(GO:0030674) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 27.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 6.9 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 3.3 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 2.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 8.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 2.1 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 1.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 2.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 3.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 5.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 2.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.7 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 28.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.5 | 15.0 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 5.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 3.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 7.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 7.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 1.9 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 3.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 2.6 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 5.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 2.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 3.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.4 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 2.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 2.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 2.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 6.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 2.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.2 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 1.3 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 3.9 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.3 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |