Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Tbx20
Z-value: 0.52
Transcription factors associated with Tbx20
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx20
|
ENSMUSG00000031965.16 | Tbx20 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx20 | mm39_v1_chr9_-_24685572_24685599 | 0.01 | 9.4e-01 | Click! |
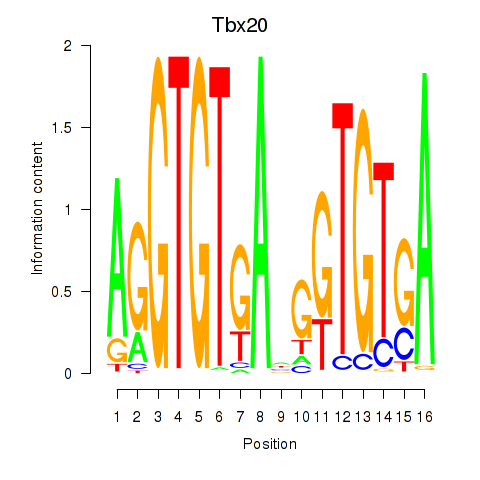
Activity profile of Tbx20 motif
Sorted Z-values of Tbx20 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx20
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_106660470 | 9.97 |
ENSMUST00000034368.8
|
Ctrl
|
chymotrypsin-like |
| chr5_-_53864874 | 1.13 |
ENSMUST00000031093.5
|
Cckar
|
cholecystokinin A receptor |
| chr8_-_23184070 | 0.44 |
ENSMUST00000131767.2
|
Ikbkb
|
inhibitor of kappaB kinase beta |
| chr2_+_122129379 | 0.25 |
ENSMUST00000028656.2
|
Duoxa2
|
dual oxidase maturation factor 2 |
| chr9_+_44410417 | 0.25 |
ENSMUST00000074989.7
ENSMUST00000218913.2 |
Bcl9l
|
B cell CLL/lymphoma 9-like |
| chr11_-_21321144 | 0.23 |
ENSMUST00000060895.6
|
Ugp2
|
UDP-glucose pyrophosphorylase 2 |
| chr1_-_136877277 | 0.22 |
ENSMUST00000168126.7
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr1_-_136876902 | 0.19 |
ENSMUST00000195428.2
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr7_+_114344920 | 0.19 |
ENSMUST00000136645.8
ENSMUST00000169913.8 |
Insc
|
INSC spindle orientation adaptor protein |
| chr7_-_105436775 | 0.18 |
ENSMUST00000078482.13
|
Dchs1
|
dachsous cadherin related 1 |
| chr17_-_13087012 | 0.18 |
ENSMUST00000089015.10
|
Mas1
|
MAS1 oncogene |
| chrX_+_98086187 | 0.18 |
ENSMUST00000036606.14
|
Stard8
|
START domain containing 8 |
| chr11_-_51748450 | 0.17 |
ENSMUST00000020655.14
ENSMUST00000109090.8 |
Jade2
|
jade family PHD finger 2 |
| chr2_-_122128930 | 0.13 |
ENSMUST00000237546.2
|
Duox2
|
dual oxidase 2 |
| chr11_+_98239230 | 0.12 |
ENSMUST00000078694.13
|
Ppp1r1b
|
protein phosphatase 1, regulatory inhibitor subunit 1B |
| chr11_-_23720953 | 0.11 |
ENSMUST00000102864.5
|
Rel
|
reticuloendotheliosis oncogene |
| chr8_-_129791969 | 0.08 |
ENSMUST00000125112.8
ENSMUST00000108747.3 ENSMUST00000214889.2 ENSMUST00000095158.11 |
Ccdc7a
|
coiled-coil domain containing 7A |
| chr9_-_72946928 | 0.08 |
ENSMUST00000184389.8
|
Pigb
|
phosphatidylinositol glycan anchor biosynthesis, class B |
| chr8_+_129793610 | 0.06 |
ENSMUST00000108745.8
|
Ccdc7b
|
coiled-coil domain containing 7B |
| chr8_+_129793635 | 0.06 |
ENSMUST00000026912.8
|
Ccdc7b
|
coiled-coil domain containing 7B |
| chr10_+_75160692 | 0.06 |
ENSMUST00000217703.2
|
Adora2a
|
adenosine A2a receptor |
| chr7_-_104577264 | 0.06 |
ENSMUST00000215359.2
|
Olfr668
|
olfactory receptor 668 |
| chr2_+_54326329 | 0.05 |
ENSMUST00000112636.8
ENSMUST00000112635.8 ENSMUST00000112634.8 |
Galnt13
|
polypeptide N-acetylgalactosaminyltransferase 13 |
| chr4_+_152200770 | 0.04 |
ENSMUST00000035275.8
|
Tnfrsf25
|
tumor necrosis factor receptor superfamily, member 25 |
| chr17_-_29456750 | 0.04 |
ENSMUST00000137727.3
|
Cpne5
|
copine V |
| chr19_+_5497575 | 0.02 |
ENSMUST00000025850.7
ENSMUST00000236774.2 |
Fosl1
|
fos-like antigen 1 |
| chr1_+_24216691 | 0.01 |
ENSMUST00000054588.15
|
Col9a1
|
collagen, type IX, alpha 1 |
| chr12_+_51395154 | 0.01 |
ENSMUST00000121521.2
|
G2e3
|
G2/M-phase specific E3 ubiquitin ligase |
| chrY_+_7722941 | 0.01 |
ENSMUST00000189007.2
|
Gm28576
|
predicted gene 28576 |
| chrY_+_7253787 | 0.01 |
ENSMUST00000186194.2
|
Gm28998
|
predicted gene 28998 |
| chr6_+_15196950 | 0.01 |
ENSMUST00000140557.8
ENSMUST00000131414.8 ENSMUST00000115469.8 |
Foxp2
|
forkhead box P2 |
| chr9_+_35580941 | 0.00 |
ENSMUST00000217565.2
|
Pate2
|
prostate and testis expressed 2 |
| chr7_-_45750050 | 0.00 |
ENSMUST00000209291.2
|
Kcnj11
|
potassium inwardly rectifying channel, subfamily J, member 11 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0090274 | reduction of food intake in response to dietary excess(GO:0002023) positive regulation of somatostatin secretion(GO:0090274) |
| 0.1 | 0.2 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.1 | 0.4 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.2 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.4 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.3 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.1 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.2 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.0 | 10.0 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 0.2 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.2 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |