Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
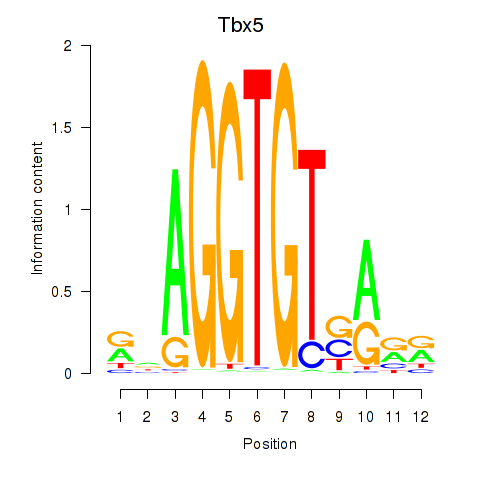
Results for Tbx5
Z-value: 2.01
Transcription factors associated with Tbx5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx5
|
ENSMUSG00000018263.12 | Tbx5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx5 | mm39_v1_chr5_+_119970733_119970780 | 0.09 | 5.9e-01 | Click! |
Activity profile of Tbx5 motif
Sorted Z-values of Tbx5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_106660470 | 15.12 |
ENSMUST00000034368.8
|
Ctrl
|
chymotrypsin-like |
| chr3_-_20329823 | 12.20 |
ENSMUST00000011607.6
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr6_+_30639217 | 11.85 |
ENSMUST00000031806.10
|
Cpa1
|
carboxypeptidase A1, pancreatic |
| chr7_-_126651847 | 7.65 |
ENSMUST00000205424.2
|
Zg16
|
zymogen granule protein 16 |
| chr15_-_100583044 | 7.28 |
ENSMUST00000230312.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr14_+_80237691 | 7.17 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr3_-_100396635 | 7.06 |
ENSMUST00000061455.9
|
Tent5c
|
terminal nucleotidyltransferase 5C |
| chr10_+_43455919 | 6.78 |
ENSMUST00000214476.2
|
Cd24a
|
CD24a antigen |
| chr1_-_132318039 | 6.74 |
ENSMUST00000132435.2
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr15_-_103163860 | 6.55 |
ENSMUST00000075192.13
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr14_-_70855980 | 6.36 |
ENSMUST00000228001.2
|
Dmtn
|
dematin actin binding protein |
| chr7_-_142220553 | 6.32 |
ENSMUST00000105935.8
|
Igf2
|
insulin-like growth factor 2 |
| chr10_+_75402090 | 6.08 |
ENSMUST00000129232.8
ENSMUST00000143792.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr8_+_23525101 | 5.74 |
ENSMUST00000117662.8
ENSMUST00000117296.8 ENSMUST00000141784.9 |
Ank1
|
ankyrin 1, erythroid |
| chr2_-_32277773 | 5.10 |
ENSMUST00000050785.14
|
Lcn2
|
lipocalin 2 |
| chr2_+_103800553 | 5.04 |
ENSMUST00000111140.3
ENSMUST00000111139.3 |
Lmo2
|
LIM domain only 2 |
| chr15_+_73620213 | 4.99 |
ENSMUST00000053232.8
|
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr2_+_103800459 | 4.81 |
ENSMUST00000111143.8
ENSMUST00000138815.2 |
Lmo2
|
LIM domain only 2 |
| chr11_+_70529944 | 4.76 |
ENSMUST00000055184.7
ENSMUST00000108551.3 |
Gp1ba
|
glycoprotein 1b, alpha polypeptide |
| chr7_-_44888220 | 4.56 |
ENSMUST00000210372.2
ENSMUST00000209779.2 ENSMUST00000098461.10 ENSMUST00000211373.2 |
Cd37
|
CD37 antigen |
| chr2_-_32278245 | 4.56 |
ENSMUST00000192241.2
|
Lcn2
|
lipocalin 2 |
| chr5_-_148336711 | 4.54 |
ENSMUST00000048116.15
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr9_-_95727267 | 4.51 |
ENSMUST00000093800.9
|
Pls1
|
plastin 1 (I-isoform) |
| chr15_-_89310060 | 4.38 |
ENSMUST00000109313.9
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr11_+_53410552 | 4.00 |
ENSMUST00000108987.8
ENSMUST00000121334.8 ENSMUST00000117061.8 |
Septin8
|
septin 8 |
| chr4_+_114916703 | 3.86 |
ENSMUST00000162489.2
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr7_-_44888532 | 3.85 |
ENSMUST00000033063.15
|
Cd37
|
CD37 antigen |
| chr7_+_28682253 | 3.85 |
ENSMUST00000085835.8
|
Map4k1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr11_-_87766350 | 3.77 |
ENSMUST00000049768.4
|
Epx
|
eosinophil peroxidase |
| chr14_-_56322654 | 3.71 |
ENSMUST00000015594.9
|
Mcpt8
|
mast cell protease 8 |
| chr15_-_82917495 | 3.53 |
ENSMUST00000231165.2
|
Nfam1
|
Nfat activating molecule with ITAM motif 1 |
| chrX_-_36253309 | 3.52 |
ENSMUST00000060474.14
ENSMUST00000053456.11 ENSMUST00000115239.10 |
Septin6
|
septin 6 |
| chr10_+_60182630 | 3.45 |
ENSMUST00000020301.14
ENSMUST00000105460.8 ENSMUST00000170507.8 |
Vsir
|
V-set immunoregulatory receptor |
| chr2_-_181333597 | 3.43 |
ENSMUST00000108778.8
ENSMUST00000165416.8 |
Rgs19
|
regulator of G-protein signaling 19 |
| chr7_+_81508741 | 3.32 |
ENSMUST00000041890.8
|
Tm6sf1
|
transmembrane 6 superfamily member 1 |
| chr6_-_49191891 | 3.30 |
ENSMUST00000031838.9
|
Igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr17_+_47816137 | 3.18 |
ENSMUST00000182935.8
ENSMUST00000182506.8 |
Ccnd3
|
cyclin D3 |
| chr17_+_47816074 | 3.17 |
ENSMUST00000183177.8
ENSMUST00000182848.8 |
Ccnd3
|
cyclin D3 |
| chr3_+_108291145 | 3.14 |
ENSMUST00000090561.10
ENSMUST00000102629.8 ENSMUST00000128089.2 |
Psrc1
|
proline/serine-rich coiled-coil 1 |
| chr17_+_47815968 | 3.14 |
ENSMUST00000182129.8
ENSMUST00000171031.8 |
Ccnd3
|
cyclin D3 |
| chr17_-_28736483 | 2.99 |
ENSMUST00000114792.8
ENSMUST00000177939.8 |
Fkbp5
|
FK506 binding protein 5 |
| chr11_+_68986043 | 2.96 |
ENSMUST00000101004.9
|
Per1
|
period circadian clock 1 |
| chr17_+_47908025 | 2.92 |
ENSMUST00000183206.2
|
Ccnd3
|
cyclin D3 |
| chr11_-_46203047 | 2.91 |
ENSMUST00000129474.2
ENSMUST00000093166.11 ENSMUST00000165599.9 |
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr19_+_3372296 | 2.85 |
ENSMUST00000237938.2
|
Cpt1a
|
carnitine palmitoyltransferase 1a, liver |
| chr11_+_115044966 | 2.82 |
ENSMUST00000021076.6
|
Rab37
|
RAB37, member RAS oncogene family |
| chr6_+_35229628 | 2.81 |
ENSMUST00000130875.8
|
1810058I24Rik
|
RIKEN cDNA 1810058I24 gene |
| chr15_+_79578141 | 2.79 |
ENSMUST00000230898.2
ENSMUST00000229046.2 |
Gtpbp1
|
GTP binding protein 1 |
| chr7_+_43086432 | 2.67 |
ENSMUST00000070518.4
|
Nkg7
|
natural killer cell group 7 sequence |
| chr7_-_44888465 | 2.66 |
ENSMUST00000210078.2
|
Cd37
|
CD37 antigen |
| chrX_+_99669343 | 2.63 |
ENSMUST00000048962.4
|
Kif4
|
kinesin family member 4 |
| chr17_+_47816042 | 2.61 |
ENSMUST00000183044.8
ENSMUST00000037333.17 |
Ccnd3
|
cyclin D3 |
| chr7_+_140711181 | 2.57 |
ENSMUST00000026568.10
|
Ptdss2
|
phosphatidylserine synthase 2 |
| chr12_-_114252202 | 2.49 |
ENSMUST00000195124.6
ENSMUST00000103481.3 |
Ighv3-6
|
immunoglobulin heavy variable 3-6 |
| chr5_-_22549688 | 2.47 |
ENSMUST00000062372.14
ENSMUST00000161356.8 |
Reln
|
reelin |
| chr11_+_78079243 | 2.44 |
ENSMUST00000002128.14
ENSMUST00000150941.8 |
Rab34
|
RAB34, member RAS oncogene family |
| chr10_+_80692948 | 2.39 |
ENSMUST00000220091.2
|
Sppl2b
|
signal peptide peptidase like 2B |
| chr6_+_35229589 | 2.34 |
ENSMUST00000152147.8
|
1810058I24Rik
|
RIKEN cDNA 1810058I24 gene |
| chr9_+_72952115 | 2.33 |
ENSMUST00000184146.8
ENSMUST00000034722.5 |
Rab27a
|
RAB27A, member RAS oncogene family |
| chr6_+_4504814 | 2.31 |
ENSMUST00000141483.8
|
Col1a2
|
collagen, type I, alpha 2 |
| chr15_-_82128538 | 2.30 |
ENSMUST00000229747.2
ENSMUST00000230408.2 |
Cenpm
|
centromere protein M |
| chr17_+_48571298 | 2.29 |
ENSMUST00000059873.14
ENSMUST00000154335.8 ENSMUST00000136272.8 ENSMUST00000125426.8 ENSMUST00000153420.2 |
Treml4
|
triggering receptor expressed on myeloid cells-like 4 |
| chr3_-_92493507 | 2.21 |
ENSMUST00000194965.6
|
Smcp
|
sperm mitochondria-associated cysteine-rich protein |
| chr14_-_51295099 | 2.19 |
ENSMUST00000227764.2
|
Rnase12
|
ribonuclease, RNase A family, 12 (non-active) |
| chr19_-_10282218 | 2.18 |
ENSMUST00000039327.11
|
Dagla
|
diacylglycerol lipase, alpha |
| chr7_+_43086554 | 2.17 |
ENSMUST00000206741.2
|
Nkg7
|
natural killer cell group 7 sequence |
| chr15_+_79231720 | 2.15 |
ENSMUST00000096350.11
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr3_+_90173813 | 2.15 |
ENSMUST00000098914.10
|
Dennd4b
|
DENN/MADD domain containing 4B |
| chr9_+_37439367 | 2.11 |
ENSMUST00000002011.14
|
Esam
|
endothelial cell-specific adhesion molecule |
| chr11_-_100305654 | 2.03 |
ENSMUST00000066489.13
|
P3h4
|
prolyl 3-hydroxylase family member 4 (non-enzymatic) |
| chr19_+_38085510 | 2.02 |
ENSMUST00000067098.8
|
Ffar4
|
free fatty acid receptor 4 |
| chr4_-_43045685 | 1.99 |
ENSMUST00000107956.8
ENSMUST00000107957.8 |
Fam214b
|
family with sequence similarity 214, member B |
| chrX_-_48886577 | 1.97 |
ENSMUST00000033442.14
ENSMUST00000114891.2 |
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr12_-_113823290 | 1.92 |
ENSMUST00000103459.5
|
Ighv5-17
|
immunoglobulin heavy variable 5-17 |
| chr11_+_69792642 | 1.92 |
ENSMUST00000177138.8
ENSMUST00000108617.10 ENSMUST00000177476.8 ENSMUST00000061837.11 |
Neurl4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr1_+_91294133 | 1.86 |
ENSMUST00000086861.12
|
Erfe
|
erythroferrone |
| chr7_-_45044606 | 1.85 |
ENSMUST00000209858.2
|
Snrnp70
|
small nuclear ribonucleoprotein 70 (U1) |
| chr15_+_79230777 | 1.81 |
ENSMUST00000229130.2
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr18_-_55123153 | 1.80 |
ENSMUST00000064763.7
|
Zfp608
|
zinc finger protein 608 |
| chr1_+_39407183 | 1.79 |
ENSMUST00000195123.6
|
Rpl31
|
ribosomal protein L31 |
| chr7_-_25488060 | 1.76 |
ENSMUST00000002677.11
ENSMUST00000085948.11 |
Axl
|
AXL receptor tyrosine kinase |
| chr7_-_141023902 | 1.76 |
ENSMUST00000026580.12
|
Pidd1
|
p53 induced death domain protein 1 |
| chr9_-_106762818 | 1.69 |
ENSMUST00000185707.2
|
Rbm15b
|
RNA binding motif protein 15B |
| chr4_+_132495636 | 1.68 |
ENSMUST00000102561.11
|
Rpa2
|
replication protein A2 |
| chr4_-_117740624 | 1.64 |
ENSMUST00000030266.12
|
B4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr19_+_54033681 | 1.63 |
ENSMUST00000237285.2
|
Adra2a
|
adrenergic receptor, alpha 2a |
| chr11_+_78079562 | 1.62 |
ENSMUST00000108322.9
|
Rab34
|
RAB34, member RAS oncogene family |
| chr7_+_89779564 | 1.62 |
ENSMUST00000208742.2
ENSMUST00000049537.9 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr1_-_167112784 | 1.60 |
ENSMUST00000053686.9
|
Uck2
|
uridine-cytidine kinase 2 |
| chr6_-_40562700 | 1.58 |
ENSMUST00000177178.2
ENSMUST00000129948.9 ENSMUST00000101491.11 |
Clec5a
|
C-type lectin domain family 5, member a |
| chr7_+_24967094 | 1.57 |
ENSMUST00000169266.8
|
Cic
|
capicua transcriptional repressor |
| chr9_+_108820846 | 1.57 |
ENSMUST00000198140.5
ENSMUST00000051873.15 |
Pfkfb4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr7_-_24771717 | 1.55 |
ENSMUST00000003468.10
|
Grik5
|
glutamate receptor, ionotropic, kainate 5 (gamma 2) |
| chr10_+_122514669 | 1.46 |
ENSMUST00000161487.8
ENSMUST00000067918.12 |
Ppm1h
|
protein phosphatase 1H (PP2C domain containing) |
| chr7_-_35096133 | 1.41 |
ENSMUST00000154597.2
ENSMUST00000032704.12 |
Faap24
|
Fanconi anemia core complex associated protein 24 |
| chr6_+_17307639 | 1.40 |
ENSMUST00000115453.2
|
Cav1
|
caveolin 1, caveolae protein |
| chr8_-_85413707 | 1.39 |
ENSMUST00000238301.2
|
Nacc1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr11_+_96820220 | 1.37 |
ENSMUST00000062172.6
|
Prr15l
|
proline rich 15-like |
| chr1_+_132405099 | 1.36 |
ENSMUST00000190825.7
ENSMUST00000190997.7 ENSMUST00000187505.7 ENSMUST00000027700.15 ENSMUST00000188575.7 |
Rbbp5
|
retinoblastoma binding protein 5, histone lysine methyltransferase complex subunit |
| chr15_-_77653979 | 1.35 |
ENSMUST00000229259.2
|
Myh9
|
myosin, heavy polypeptide 9, non-muscle |
| chr14_-_70414236 | 1.34 |
ENSMUST00000153735.8
|
Pdlim2
|
PDZ and LIM domain 2 |
| chrX_+_168468186 | 1.32 |
ENSMUST00000112107.8
ENSMUST00000112104.8 |
Mid1
|
midline 1 |
| chr12_-_113700190 | 1.32 |
ENSMUST00000103452.3
ENSMUST00000192264.2 |
Ighv5-9-1
|
immunoglobulin heavy variable 5-9-1 |
| chr12_-_113589576 | 1.31 |
ENSMUST00000103446.2
|
Ighv5-6
|
immunoglobulin heavy variable 5-6 |
| chr2_-_44817173 | 1.31 |
ENSMUST00000130991.8
|
Gtdc1
|
glycosyltransferase-like domain containing 1 |
| chrX_+_158623460 | 1.30 |
ENSMUST00000112451.8
ENSMUST00000112453.9 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr16_+_31247562 | 1.30 |
ENSMUST00000115227.10
|
Bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr4_-_132125510 | 1.29 |
ENSMUST00000136711.2
ENSMUST00000084249.11 |
Phactr4
|
phosphatase and actin regulator 4 |
| chr12_-_111638722 | 1.27 |
ENSMUST00000001304.9
|
Ckb
|
creatine kinase, brain |
| chr8_+_121264161 | 1.24 |
ENSMUST00000118136.2
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr11_+_50101717 | 1.23 |
ENSMUST00000147468.8
|
Mgat4b
|
mannoside acetylglucosaminyltransferase 4, isoenzyme B |
| chr17_+_32255071 | 1.22 |
ENSMUST00000081339.13
|
Rrp1b
|
ribosomal RNA processing 1B |
| chr9_-_62444318 | 1.20 |
ENSMUST00000048043.12
|
Coro2b
|
coronin, actin binding protein, 2B |
| chr4_-_149858694 | 1.20 |
ENSMUST00000105686.3
|
Slc25a33
|
solute carrier family 25, member 33 |
| chr9_+_69902697 | 1.19 |
ENSMUST00000165389.8
|
Bnip2
|
BCL2/adenovirus E1B interacting protein 2 |
| chr14_+_30973407 | 1.17 |
ENSMUST00000022458.11
|
Bap1
|
Brca1 associated protein 1 |
| chr8_+_121262528 | 1.16 |
ENSMUST00000120493.8
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr4_+_154255187 | 1.15 |
ENSMUST00000030897.15
|
Megf6
|
multiple EGF-like-domains 6 |
| chr4_-_133746138 | 1.15 |
ENSMUST00000051674.3
|
Lin28a
|
lin-28 homolog A (C. elegans) |
| chr3_+_123240562 | 1.14 |
ENSMUST00000029603.10
|
Prss12
|
protease, serine 12 neurotrypsin (motopsin) |
| chr9_-_122123475 | 1.13 |
ENSMUST00000042546.4
|
Ano10
|
anoctamin 10 |
| chr18_+_82932747 | 1.12 |
ENSMUST00000071233.7
|
Zfp516
|
zinc finger protein 516 |
| chr19_-_4861536 | 1.12 |
ENSMUST00000179909.8
ENSMUST00000172000.3 ENSMUST00000180008.2 ENSMUST00000113793.4 ENSMUST00000006625.8 |
Gm21992
Rbm14
|
predicted gene 21992 RNA binding motif protein 14 |
| chr4_-_116982804 | 1.12 |
ENSMUST00000183310.2
|
Btbd19
|
BTB (POZ) domain containing 19 |
| chr11_+_83553400 | 1.10 |
ENSMUST00000019074.4
|
Ccl4
|
chemokine (C-C motif) ligand 4 |
| chr5_-_18093739 | 1.09 |
ENSMUST00000169095.6
ENSMUST00000197574.2 |
Cd36
|
CD36 molecule |
| chr9_-_122123409 | 1.07 |
ENSMUST00000214409.2
|
Ano10
|
anoctamin 10 |
| chr17_+_28910714 | 1.07 |
ENSMUST00000233250.2
|
Mapk14
|
mitogen-activated protein kinase 14 |
| chr18_+_23885390 | 1.07 |
ENSMUST00000170802.8
ENSMUST00000155708.8 ENSMUST00000118826.9 |
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr8_+_106245368 | 1.05 |
ENSMUST00000034363.7
|
Hsd11b2
|
hydroxysteroid 11-beta dehydrogenase 2 |
| chr3_+_96537235 | 1.00 |
ENSMUST00000048915.11
ENSMUST00000196456.5 ENSMUST00000198027.5 |
Rbm8a
|
RNA binding motif protein 8a |
| chr7_+_25380583 | 0.99 |
ENSMUST00000108403.4
|
B9d2
|
B9 protein domain 2 |
| chrX_+_133195974 | 0.99 |
ENSMUST00000037687.8
|
Tmem35a
|
transmembrane protein 35A |
| chr7_+_35096470 | 0.99 |
ENSMUST00000079414.12
|
Cep89
|
centrosomal protein 89 |
| chr5_-_100867520 | 0.98 |
ENSMUST00000112908.2
ENSMUST00000045617.15 |
Hpse
|
heparanase |
| chrX_-_73009933 | 0.97 |
ENSMUST00000114372.3
ENSMUST00000033761.13 |
Hcfc1
|
host cell factor C1 |
| chr3_+_96537484 | 0.94 |
ENSMUST00000200647.2
|
Rbm8a
|
RNA binding motif protein 8a |
| chr19_+_38384428 | 0.93 |
ENSMUST00000054098.4
|
Slc35g1
|
solute carrier family 35, member G1 |
| chr9_-_122123427 | 0.92 |
ENSMUST00000216670.2
|
Ano10
|
anoctamin 10 |
| chr11_+_6339061 | 0.91 |
ENSMUST00000109787.8
|
Zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr19_-_46315543 | 0.91 |
ENSMUST00000223917.2
ENSMUST00000224447.2 ENSMUST00000041391.5 ENSMUST00000096029.12 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr7_+_92426772 | 0.90 |
ENSMUST00000208945.2
|
Rab30
|
RAB30, member RAS oncogene family |
| chr15_-_99355623 | 0.88 |
ENSMUST00000023747.14
|
Nckap5l
|
NCK-associated protein 5-like |
| chr6_-_70237939 | 0.88 |
ENSMUST00000103386.3
|
Igkv6-23
|
immunoglobulin kappa variable 6-23 |
| chr11_-_3321307 | 0.88 |
ENSMUST00000101640.10
ENSMUST00000101642.10 |
Limk2
|
LIM motif-containing protein kinase 2 |
| chr4_+_132291369 | 0.88 |
ENSMUST00000070690.8
|
Ptafr
|
platelet-activating factor receptor |
| chr6_-_113320858 | 0.88 |
ENSMUST00000155543.2
ENSMUST00000032409.15 |
Camk1
|
calcium/calmodulin-dependent protein kinase I |
| chr12_-_113666198 | 0.88 |
ENSMUST00000103450.4
|
Ighv5-12
|
immunoglobulin heavy variable 5-12 |
| chr5_-_148336574 | 0.87 |
ENSMUST00000202457.4
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chrX_+_73356597 | 0.87 |
ENSMUST00000114160.2
|
Fam50a
|
family with sequence similarity 50, member A |
| chr9_+_86625694 | 0.85 |
ENSMUST00000179574.2
ENSMUST00000036426.13 |
Prss35
|
protease, serine 35 |
| chr4_+_154954042 | 0.83 |
ENSMUST00000079269.14
ENSMUST00000163732.8 ENSMUST00000080559.13 |
Mmel1
|
membrane metallo-endopeptidase-like 1 |
| chr2_-_131987008 | 0.82 |
ENSMUST00000028815.15
|
Slc23a2
|
solute carrier family 23 (nucleobase transporters), member 2 |
| chr9_-_116004386 | 0.81 |
ENSMUST00000035014.8
|
Tgfbr2
|
transforming growth factor, beta receptor II |
| chr11_+_20597149 | 0.80 |
ENSMUST00000109585.2
|
Sertad2
|
SERTA domain containing 2 |
| chr6_-_136638926 | 0.79 |
ENSMUST00000032336.7
|
Plbd1
|
phospholipase B domain containing 1 |
| chr6_-_49240944 | 0.79 |
ENSMUST00000204189.3
ENSMUST00000031841.9 |
Tra2a
|
transformer 2 alpha |
| chr13_+_49835667 | 0.79 |
ENSMUST00000172254.3
|
Iars
|
isoleucine-tRNA synthetase |
| chr9_-_62888156 | 0.78 |
ENSMUST00000098651.6
ENSMUST00000214830.2 |
Pias1
|
protein inhibitor of activated STAT 1 |
| chr18_+_36926929 | 0.78 |
ENSMUST00000001419.10
|
Zmat2
|
zinc finger, matrin type 2 |
| chr17_+_28910393 | 0.78 |
ENSMUST00000124886.9
ENSMUST00000114758.9 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chr19_-_41373526 | 0.77 |
ENSMUST00000059672.9
|
Pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr6_+_17307272 | 0.77 |
ENSMUST00000115454.2
|
Cav1
|
caveolin 1, caveolae protein |
| chr10_-_90918566 | 0.76 |
ENSMUST00000162618.8
ENSMUST00000020157.13 ENSMUST00000160788.2 |
Apaf1
|
apoptotic peptidase activating factor 1 |
| chr2_+_24076481 | 0.76 |
ENSMUST00000057567.3
|
Il1f9
|
interleukin 1 family, member 9 |
| chr1_-_74640504 | 0.76 |
ENSMUST00000136078.2
ENSMUST00000132081.2 ENSMUST00000113721.8 ENSMUST00000027357.12 |
Rnf25
|
ring finger protein 25 |
| chr19_-_10581622 | 0.76 |
ENSMUST00000037678.7
|
Tkfc
|
triokinase, FMN cyclase |
| chr6_+_113284098 | 0.76 |
ENSMUST00000113122.8
ENSMUST00000204198.3 ENSMUST00000113121.8 ENSMUST00000113119.8 ENSMUST00000113117.4 ENSMUST00000204626.3 ENSMUST00000203577.2 |
Brpf1
|
bromodomain and PHD finger containing, 1 |
| chr3_-_95264467 | 0.76 |
ENSMUST00000107171.10
ENSMUST00000015841.12 ENSMUST00000107170.3 |
Setdb1
|
SET domain, bifurcated 1 |
| chr16_+_93480061 | 0.76 |
ENSMUST00000039620.7
|
Cbr3
|
carbonyl reductase 3 |
| chr11_+_77923172 | 0.74 |
ENSMUST00000122342.2
ENSMUST00000092881.4 |
Dhrs13
|
dehydrogenase/reductase (SDR family) member 13 |
| chrX_-_7606445 | 0.73 |
ENSMUST00000128289.8
|
Ccdc120
|
coiled-coil domain containing 120 |
| chr7_-_3669882 | 0.72 |
ENSMUST00000145034.3
|
Tmc4
|
transmembrane channel-like gene family 4 |
| chr3_+_126390600 | 0.70 |
ENSMUST00000106402.8
ENSMUST00000106399.8 ENSMUST00000066466.13 ENSMUST00000163226.8 ENSMUST00000199300.5 |
Camk2d
|
calcium/calmodulin-dependent protein kinase II, delta |
| chr1_-_131172903 | 0.70 |
ENSMUST00000027688.15
ENSMUST00000112442.2 |
Rassf5
|
Ras association (RalGDS/AF-6) domain family member 5 |
| chr3_-_126792056 | 0.70 |
ENSMUST00000044443.15
|
Ank2
|
ankyrin 2, brain |
| chr3_-_90373165 | 0.69 |
ENSMUST00000029540.13
|
Npr1
|
natriuretic peptide receptor 1 |
| chr7_-_140697719 | 0.69 |
ENSMUST00000067836.9
|
Ano9
|
anoctamin 9 |
| chr10_-_23977810 | 0.67 |
ENSMUST00000170267.3
|
Taar8c
|
trace amine-associated receptor 8C |
| chr12_-_113733922 | 0.67 |
ENSMUST00000180013.3
|
Ighv2-9-1
|
immunoglobulin heavy variable 2-9-1 |
| chr12_-_113790741 | 0.67 |
ENSMUST00000103457.3
ENSMUST00000192877.2 |
Ighv5-15
|
immunoglobulin heavy variable 5-15 |
| chr2_-_121287174 | 0.66 |
ENSMUST00000110613.9
ENSMUST00000056312.10 |
Serinc4
|
serine incorporator 4 |
| chr16_-_18107046 | 0.65 |
ENSMUST00000232424.2
ENSMUST00000009321.11 |
Dgcr8
|
DGCR8, microprocessor complex subunit |
| chr7_+_125151432 | 0.65 |
ENSMUST00000206217.2
ENSMUST00000205985.2 |
Il4ra
|
interleukin 4 receptor, alpha |
| chr2_-_25136857 | 0.65 |
ENSMUST00000228627.3
|
Ndor1
|
NADPH dependent diflavin oxidoreductase 1 |
| chrX_+_20230720 | 0.63 |
ENSMUST00000033372.13
ENSMUST00000115391.8 ENSMUST00000115387.8 |
Rp2
|
retinitis pigmentosa 2 homolog |
| chr11_-_96834771 | 0.63 |
ENSMUST00000107629.2
ENSMUST00000018803.12 |
Pnpo
|
pyridoxine 5'-phosphate oxidase |
| chr2_-_126985226 | 0.63 |
ENSMUST00000110386.2
ENSMUST00000132773.2 |
Itpripl1
|
inositol 1,4,5-triphosphate receptor interacting protein-like 1 |
| chr2_-_91274967 | 0.63 |
ENSMUST00000064652.14
ENSMUST00000102594.11 ENSMUST00000094835.9 |
1110051M20Rik
|
RIKEN cDNA 1110051M20 gene |
| chr8_+_55024446 | 0.63 |
ENSMUST00000239166.2
ENSMUST00000239106.2 ENSMUST00000239152.2 |
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr7_-_28297565 | 0.62 |
ENSMUST00000040531.9
ENSMUST00000108283.8 |
Samd4b
Pak4
|
sterile alpha motif domain containing 4B p21 (RAC1) activated kinase 4 |
| chr11_-_70111796 | 0.62 |
ENSMUST00000060010.3
ENSMUST00000190533.2 |
Slc16a13
|
solute carrier family 16 (monocarboxylic acid transporters), member 13 |
| chr10_+_56253418 | 0.62 |
ENSMUST00000068581.9
ENSMUST00000217789.2 |
Gja1
|
gap junction protein, alpha 1 |
| chr1_+_74448535 | 0.59 |
ENSMUST00000027366.13
|
Vil1
|
villin 1 |
| chr4_+_62398262 | 0.58 |
ENSMUST00000030088.12
ENSMUST00000107449.4 |
Bspry
|
B-box and SPRY domain containing |
| chr3_-_95214102 | 0.57 |
ENSMUST00000107183.8
ENSMUST00000164406.8 ENSMUST00000123365.2 |
Anxa9
|
annexin A9 |
| chr11_-_69791712 | 0.57 |
ENSMUST00000108621.9
ENSMUST00000100969.9 |
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr3_+_32762656 | 0.56 |
ENSMUST00000029214.14
|
Actl6a
|
actin-like 6A |
| chr2_-_119039247 | 0.55 |
ENSMUST00000038439.4
|
Dnajc17
|
DnaJ heat shock protein family (Hsp40) member C17 |
| chr11_+_77923100 | 0.55 |
ENSMUST00000021187.12
|
Dhrs13
|
dehydrogenase/reductase (SDR family) member 13 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.7 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 2.3 | 6.8 | GO:0034118 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 1.3 | 3.9 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 1.3 | 5.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 1.3 | 3.8 | GO:0002215 | defense response to nematode(GO:0002215) |
| 1.1 | 6.4 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 1.0 | 7.3 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 1.0 | 11.1 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.8 | 4.1 | GO:1905169 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.8 | 6.1 | GO:1901748 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.6 | 2.5 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.6 | 9.8 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.6 | 14.5 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.5 | 2.2 | GO:0099541 | diacylglycerol catabolic process(GO:0046340) trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.5 | 1.6 | GO:0010138 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) CMP metabolic process(GO:0046035) |
| 0.5 | 6.3 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.5 | 3.0 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.5 | 3.4 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.5 | 1.5 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) |
| 0.4 | 6.7 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.4 | 0.9 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.4 | 1.3 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.4 | 2.6 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.4 | 2.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.4 | 3.5 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.4 | 1.8 | GO:0097048 | dendritic cell apoptotic process(GO:0097048) neutrophil clearance(GO:0097350) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.4 | 1.1 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.3 | 0.7 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.3 | 6.1 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.3 | 1.3 | GO:1903923 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.3 | 3.4 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.3 | 1.0 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.3 | 1.0 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of tolerance induction to self antigen(GO:0002649) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.3 | 1.3 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.3 | 5.4 | GO:0015809 | arginine transport(GO:0015809) |
| 0.3 | 2.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.3 | 2.4 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.3 | 1.2 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.3 | 2.3 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.3 | 0.8 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.3 | 1.6 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.3 | 2.4 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.3 | 1.5 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.3 | 0.8 | GO:0042376 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.2 | 1.0 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.2 | 2.6 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.2 | 1.2 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.2 | 1.6 | GO:0035625 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.2 | 1.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.2 | 0.9 | GO:1902943 | regulation of voltage-gated chloride channel activity(GO:1902941) positive regulation of voltage-gated chloride channel activity(GO:1902943) |
| 0.2 | 1.1 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.2 | 0.6 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.2 | 0.6 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.2 | 0.6 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.2 | 0.6 | GO:0010652 | positive regulation of glomerular filtration(GO:0003104) atrial ventricular junction remodeling(GO:0003294) regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.2 | 1.8 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 | 0.8 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.2 | 1.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.2 | 0.8 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.2 | 2.9 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.2 | 0.5 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.2 | 0.5 | GO:0019265 | glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) L-alanine catabolic process(GO:0042853) oxalic acid secretion(GO:0046724) |
| 0.2 | 1.6 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 2.3 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.2 | 0.5 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 | 0.4 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.1 | 1.1 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 6.2 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.1 | 0.8 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.1 | 7.5 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 1.0 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.3 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.1 | 3.4 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 0.9 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 3.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.3 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 0.1 | 1.1 | GO:2000193 | positive regulation of fatty acid transport(GO:2000193) |
| 0.1 | 0.6 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 0.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 2.2 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 0.3 | GO:0061296 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.1 | 1.6 | GO:0033033 | negative regulation of myeloid cell apoptotic process(GO:0033033) |
| 0.1 | 3.4 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 0.7 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 2.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.3 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 0.2 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.1 | 3.1 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.1 | 0.3 | GO:0046016 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) positive regulation of transcription by glucose(GO:0046016) |
| 0.1 | 3.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.5 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 1.0 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 3.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.3 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.8 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.5 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 6.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 2.6 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.2 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.7 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.1 | GO:0061723 | glycophagy(GO:0061723) |
| 0.0 | 0.1 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.0 | 0.4 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 | 2.8 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 1.6 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.4 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 | 2.0 | GO:0050710 | negative regulation of cytokine secretion(GO:0050710) |
| 0.0 | 0.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 1.6 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 3.5 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.3 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.0 | 1.5 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 1.8 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.3 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.5 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.2 | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.0 | 0.2 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 1.1 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.2 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.2 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.3 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.4 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.1 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.0 | 0.4 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 0.5 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.8 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.5 | GO:1901099 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 20.4 | GO:0006508 | proteolysis(GO:0006508) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.9 | 6.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.8 | 2.3 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.6 | 7.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.5 | 4.5 | GO:1990357 | terminal web(GO:1990357) |
| 0.4 | 11.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.4 | 2.4 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.3 | 7.2 | GO:0042581 | specific granule(GO:0042581) |
| 0.3 | 2.7 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.3 | 5.7 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.2 | 1.6 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.2 | 0.7 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.2 | 14.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 1.6 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 2.8 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.2 | 1.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.4 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 1.9 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 2.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.7 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 6.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 1.5 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 1.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.8 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 2.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 1.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 2.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 1.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.6 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.1 | 1.8 | GO:0018995 | host(GO:0018995) host cell part(GO:0033643) host cell(GO:0043657) |
| 0.1 | 7.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.3 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 3.0 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 4.3 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 1.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 1.0 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 1.9 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 7.3 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 5.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 2.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.4 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 1.0 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.3 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 3.3 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.8 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 1.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 2.1 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 2.0 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 1.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 41.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 1.7 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 6.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 4.7 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.9 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.6 | GO:0030315 | T-tubule(GO:0030315) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 7.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.8 | 24.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.6 | 5.4 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.6 | 5.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.5 | 1.6 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.5 | 6.1 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.4 | 1.3 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.4 | 1.3 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.4 | 1.6 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.4 | 1.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.4 | 2.5 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.4 | 1.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.3 | 1.0 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.3 | 1.5 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.3 | 6.8 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.3 | 4.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.3 | 0.8 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.3 | 1.6 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.3 | 1.5 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.3 | 0.8 | GO:0004371 | glycerone kinase activity(GO:0004371) FAD-AMP lyase (cyclizing) activity(GO:0034012) triokinase activity(GO:0050354) |
| 0.2 | 14.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.2 | 2.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.2 | 1.6 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.2 | 2.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 16.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 1.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 2.0 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.2 | 2.6 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.2 | 1.6 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 1.3 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.2 | 2.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 0.8 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 3.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.9 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 4.1 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 3.0 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.4 | GO:0098973 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 1.9 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 6.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 2.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.7 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 0.8 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 1.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 1.7 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 0.7 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 1.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.8 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 6.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 3.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 1.6 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.6 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.1 | 3.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 1.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 2.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.5 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.1 | 6.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.7 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 1.0 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 2.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 19.9 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.1 | 1.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.8 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 0.6 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 0.6 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 1.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 7.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.3 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 0.3 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.5 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.1 | 3.6 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 0.7 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 2.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 2.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.2 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.0 | 3.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.2 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.4 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 6.5 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.1 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.8 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 5.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.3 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.5 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 2.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 1.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.8 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.5 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.3 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.1 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.0 | 2.2 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 13.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 5.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 4.7 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 4.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 3.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 8.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 1.5 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 0.8 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 1.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 5.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 0.8 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 2.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 2.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 2.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.0 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 2.0 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 6.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.2 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.3 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.3 | 6.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 14.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.2 | 5.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 1.7 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 2.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 5.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 2.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 5.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 5.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 0.5 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 9.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 2.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 3.0 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 1.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 3.9 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.0 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.8 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 1.9 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.8 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 1.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 2.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 2.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.7 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 2.6 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.3 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 1.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |