Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Tcf7l1
Z-value: 1.19
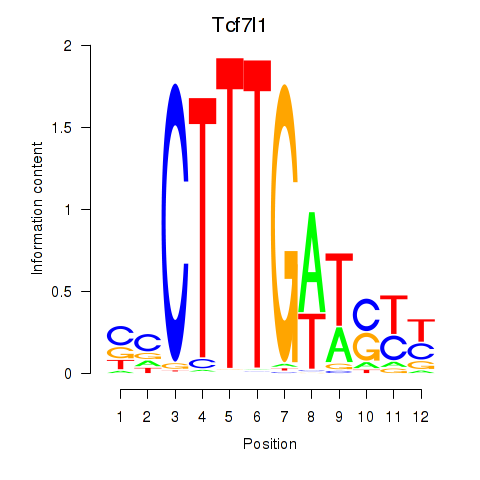
Transcription factors associated with Tcf7l1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tcf7l1
|
ENSMUSG00000055799.14 | Tcf7l1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tcf7l1 | mm39_v1_chr6_-_72765935_72766044 | 0.66 | 1.1e-05 | Click! |
Activity profile of Tcf7l1 motif
Sorted Z-values of Tcf7l1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tcf7l1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_172863688 | 3.60 |
ENSMUST00000029014.16
|
Rbm38
|
RNA binding motif protein 38 |
| chr2_+_157401998 | 2.87 |
ENSMUST00000153739.9
ENSMUST00000173595.2 ENSMUST00000109526.2 ENSMUST00000173839.2 ENSMUST00000173041.8 ENSMUST00000173793.8 ENSMUST00000172487.2 ENSMUST00000088484.6 |
Nnat
|
neuronatin |
| chr12_-_113386312 | 2.60 |
ENSMUST00000177715.8
ENSMUST00000103426.3 |
Ighm
|
immunoglobulin heavy constant mu |
| chrX_-_149595711 | 2.52 |
ENSMUST00000112697.10
|
Maged2
|
MAGE family member D2 |
| chrX_-_149595873 | 2.51 |
ENSMUST00000131241.2
ENSMUST00000147152.3 ENSMUST00000143843.8 |
Maged2
|
MAGE family member D2 |
| chr4_+_114914880 | 2.19 |
ENSMUST00000161601.8
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr10_-_75776437 | 2.11 |
ENSMUST00000219979.2
|
Gm867
|
predicted gene 867 |
| chr19_-_41790458 | 2.03 |
ENSMUST00000026150.15
ENSMUST00000163265.9 ENSMUST00000177495.2 |
Arhgap19
|
Rho GTPase activating protein 19 |
| chr2_+_172864153 | 1.98 |
ENSMUST00000173997.2
|
Rbm38
|
RNA binding motif protein 38 |
| chr13_-_55635851 | 1.89 |
ENSMUST00000109921.9
ENSMUST00000109923.9 ENSMUST00000021950.15 |
Dbn1
|
drebrin 1 |
| chr18_+_21205386 | 1.85 |
ENSMUST00000082235.5
|
Mep1b
|
meprin 1 beta |
| chr7_-_83533497 | 1.83 |
ENSMUST00000094216.5
|
Tlnrd1
|
talin rod domain containing 1 |
| chr7_-_12743720 | 1.75 |
ENSMUST00000210282.2
ENSMUST00000172240.2 ENSMUST00000051390.9 ENSMUST00000209997.2 |
Zbtb45
|
zinc finger and BTB domain containing 45 |
| chr6_+_71684846 | 1.75 |
ENSMUST00000212792.2
|
Reep1
|
receptor accessory protein 1 |
| chr8_-_4309257 | 1.75 |
ENSMUST00000053252.9
|
Ctxn1
|
cortexin 1 |
| chr9_+_98372575 | 1.68 |
ENSMUST00000035029.3
|
Rbp2
|
retinol binding protein 2, cellular |
| chr14_-_61283911 | 1.60 |
ENSMUST00000111234.10
ENSMUST00000224371.2 |
Tnfrsf19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr15_-_98851423 | 1.59 |
ENSMUST00000134214.3
|
Gm49450
|
predicted gene, 49450 |
| chr15_-_98851566 | 1.56 |
ENSMUST00000097014.7
|
Tuba1a
|
tubulin, alpha 1A |
| chr1_-_45965661 | 1.44 |
ENSMUST00000186804.2
ENSMUST00000187406.7 ENSMUST00000187420.7 |
Slc40a1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr6_-_83504471 | 1.36 |
ENSMUST00000141904.8
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr6_+_29735666 | 1.35 |
ENSMUST00000001812.5
|
Smo
|
smoothened, frizzled class receptor |
| chr11_-_86999481 | 1.32 |
ENSMUST00000051395.9
|
Prr11
|
proline rich 11 |
| chr7_-_144493560 | 1.31 |
ENSMUST00000093962.5
|
Ccnd1
|
cyclin D1 |
| chr9_+_90045219 | 1.28 |
ENSMUST00000147250.8
ENSMUST00000113060.3 |
Adamts7
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 7 |
| chr9_+_90045109 | 1.28 |
ENSMUST00000113059.8
ENSMUST00000167122.8 |
Adamts7
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 7 |
| chr5_-_148329615 | 1.26 |
ENSMUST00000138257.8
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr5_+_31350607 | 1.26 |
ENSMUST00000201535.4
|
Snx17
|
sorting nexin 17 |
| chr7_+_44866095 | 1.26 |
ENSMUST00000209437.2
|
Tead2
|
TEA domain family member 2 |
| chr7_+_78922947 | 1.22 |
ENSMUST00000037315.13
|
Abhd2
|
abhydrolase domain containing 2 |
| chr6_+_83034825 | 1.20 |
ENSMUST00000077502.5
|
Dqx1
|
DEAQ RNA-dependent ATPase |
| chr6_+_17065141 | 1.19 |
ENSMUST00000115467.11
ENSMUST00000154266.3 ENSMUST00000076654.9 |
Tes
|
testin LIM domain protein |
| chr19_+_8568618 | 1.17 |
ENSMUST00000170817.2
ENSMUST00000010251.11 |
Slc22a8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr19_-_46327024 | 1.15 |
ENSMUST00000236046.2
ENSMUST00000236980.2 ENSMUST00000235485.2 ENSMUST00000236061.2 ENSMUST00000236236.2 ENSMUST00000236768.2 ENSMUST00000236651.2 |
Cuedc2
|
CUE domain containing 2 |
| chr11_-_52174129 | 1.15 |
ENSMUST00000109071.3
|
Tcf7
|
transcription factor 7, T cell specific |
| chr2_+_13579092 | 1.14 |
ENSMUST00000193675.2
|
Vim
|
vimentin |
| chr5_+_31350566 | 1.12 |
ENSMUST00000031029.15
ENSMUST00000201679.4 |
Snx17
|
sorting nexin 17 |
| chr11_+_45946800 | 1.07 |
ENSMUST00000011400.8
|
Adam19
|
a disintegrin and metallopeptidase domain 19 (meltrin beta) |
| chr7_-_110681402 | 1.06 |
ENSMUST00000159305.2
|
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr14_+_71011744 | 1.06 |
ENSMUST00000022698.8
|
Dok2
|
docking protein 2 |
| chr11_-_53959790 | 1.06 |
ENSMUST00000018755.10
|
Pdlim4
|
PDZ and LIM domain 4 |
| chr17_+_29712008 | 1.03 |
ENSMUST00000234665.2
|
Pim1
|
proviral integration site 1 |
| chr19_-_46327071 | 1.02 |
ENSMUST00000235977.2
ENSMUST00000167861.8 ENSMUST00000051234.9 ENSMUST00000236066.2 |
Cuedc2
|
CUE domain containing 2 |
| chr6_+_38895902 | 0.98 |
ENSMUST00000003017.13
|
Tbxas1
|
thromboxane A synthase 1, platelet |
| chr6_+_71684872 | 0.97 |
ENSMUST00000212631.2
|
Reep1
|
receptor accessory protein 1 |
| chr1_-_182929025 | 0.94 |
ENSMUST00000171366.7
|
Disp1
|
dispatched RND transporter family member 1 |
| chr2_+_163500290 | 0.89 |
ENSMUST00000164399.8
ENSMUST00000064703.13 ENSMUST00000099105.9 ENSMUST00000152418.8 ENSMUST00000126182.8 ENSMUST00000131228.8 |
Pkig
|
protein kinase inhibitor, gamma |
| chr4_+_130519788 | 0.89 |
ENSMUST00000070478.4
|
Sdc3
|
syndecan 3 |
| chr11_-_53959758 | 0.89 |
ENSMUST00000093109.11
|
Pdlim4
|
PDZ and LIM domain 4 |
| chr6_+_86605146 | 0.88 |
ENSMUST00000043400.9
|
Asprv1
|
aspartic peptidase, retroviral-like 1 |
| chr8_+_22996233 | 0.88 |
ENSMUST00000210854.2
|
Slc20a2
|
solute carrier family 20, member 2 |
| chr1_-_45964730 | 0.88 |
ENSMUST00000027137.11
|
Slc40a1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr11_+_96820091 | 0.87 |
ENSMUST00000054311.6
ENSMUST00000107636.4 |
Prr15l
|
proline rich 15-like |
| chr17_+_12338161 | 0.87 |
ENSMUST00000024594.9
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr5_+_33978035 | 0.84 |
ENSMUST00000075812.11
ENSMUST00000114397.9 ENSMUST00000155880.8 |
Nsd2
|
nuclear receptor binding SET domain protein 2 |
| chr11_+_96820220 | 0.82 |
ENSMUST00000062172.6
|
Prr15l
|
proline rich 15-like |
| chr1_+_87397980 | 0.80 |
ENSMUST00000027476.6
|
Snorc
|
secondary ossification center associated regulator of chondrocyte maturation |
| chr17_-_90217868 | 0.80 |
ENSMUST00000086423.6
|
Gm10184
|
predicted pseudogene 10184 |
| chr11_+_69737437 | 0.78 |
ENSMUST00000152566.8
ENSMUST00000108633.9 |
Plscr3
|
phospholipid scramblase 3 |
| chr11_+_69737200 | 0.78 |
ENSMUST00000108632.8
|
Plscr3
|
phospholipid scramblase 3 |
| chrX_+_72527208 | 0.77 |
ENSMUST00000033741.15
ENSMUST00000169489.2 |
Bgn
|
biglycan |
| chr7_-_143102792 | 0.76 |
ENSMUST00000072727.7
ENSMUST00000207948.2 ENSMUST00000208190.2 |
Nap1l4
|
nucleosome assembly protein 1-like 4 |
| chr13_+_94219934 | 0.75 |
ENSMUST00000156071.2
|
Lhfpl2
|
lipoma HMGIC fusion partner-like 2 |
| chrX_+_168662592 | 0.75 |
ENSMUST00000112105.8
ENSMUST00000078947.12 |
Mid1
|
midline 1 |
| chr13_-_119545520 | 0.73 |
ENSMUST00000069902.13
ENSMUST00000099149.10 ENSMUST00000109204.8 |
Nnt
|
nicotinamide nucleotide transhydrogenase |
| chr7_-_117728790 | 0.73 |
ENSMUST00000206491.2
|
Arl6ip1
|
ADP-ribosylation factor-like 6 interacting protein 1 |
| chr1_+_136395673 | 0.72 |
ENSMUST00000189413.7
ENSMUST00000047817.12 |
Kif14
|
kinesin family member 14 |
| chr4_-_141391406 | 0.72 |
ENSMUST00000084203.11
|
Plekhm2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2 |
| chr2_-_164876690 | 0.71 |
ENSMUST00000122070.2
ENSMUST00000121377.8 ENSMUST00000153905.2 ENSMUST00000040381.15 |
Ncoa5
|
nuclear receptor coactivator 5 |
| chr1_+_88154727 | 0.70 |
ENSMUST00000061013.13
ENSMUST00000113130.8 |
Mroh2a
|
maestro heat-like repeat family member 2A |
| chr11_+_69737491 | 0.69 |
ENSMUST00000019605.4
|
Plscr3
|
phospholipid scramblase 3 |
| chr9_-_45896663 | 0.69 |
ENSMUST00000214179.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr7_-_141649003 | 0.67 |
ENSMUST00000039926.10
|
Dusp8
|
dual specificity phosphatase 8 |
| chr19_-_10581622 | 0.67 |
ENSMUST00000037678.7
|
Tkfc
|
triokinase, FMN cyclase |
| chr14_-_70414236 | 0.65 |
ENSMUST00000153735.8
|
Pdlim2
|
PDZ and LIM domain 2 |
| chr9_-_45896110 | 0.65 |
ENSMUST00000215060.2
ENSMUST00000213853.2 ENSMUST00000216334.2 |
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr9_+_21527462 | 0.65 |
ENSMUST00000034707.15
ENSMUST00000098948.10 |
Smarca4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr8_+_31601837 | 0.65 |
ENSMUST00000046941.8
ENSMUST00000217278.2 |
Rnf122
|
ring finger protein 122 |
| chr4_+_132366298 | 0.63 |
ENSMUST00000135299.8
ENSMUST00000020197.14 ENSMUST00000180250.8 ENSMUST00000081726.13 ENSMUST00000079157.11 |
Eya3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr17_+_36176485 | 0.62 |
ENSMUST00000127442.8
ENSMUST00000144382.8 |
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr6_-_122317484 | 0.61 |
ENSMUST00000112600.9
|
Phc1
|
polyhomeotic 1 |
| chr6_+_83114020 | 0.60 |
ENSMUST00000121093.8
|
Rtkn
|
rhotekin |
| chr12_-_114443071 | 0.59 |
ENSMUST00000103492.2
|
Ighv10-1
|
immunoglobulin heavy variable 10-1 |
| chr7_-_143102524 | 0.59 |
ENSMUST00000208093.2
ENSMUST00000209098.2 |
Nap1l4
|
nucleosome assembly protein 1-like 4 |
| chr12_+_85043083 | 0.58 |
ENSMUST00000168977.8
ENSMUST00000021670.15 |
Ylpm1
|
YLP motif containing 1 |
| chr6_-_47571901 | 0.58 |
ENSMUST00000081721.13
ENSMUST00000114618.8 ENSMUST00000114616.8 |
Ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr11_-_48836975 | 0.58 |
ENSMUST00000104958.2
|
Psme2b
|
protease (prosome, macropain) activator subunit 2B |
| chr17_+_46513666 | 0.58 |
ENSMUST00000087031.7
|
Xpo5
|
exportin 5 |
| chr13_-_119545479 | 0.57 |
ENSMUST00000223268.2
|
Nnt
|
nicotinamide nucleotide transhydrogenase |
| chr10_-_13264497 | 0.57 |
ENSMUST00000105546.8
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr9_-_65734826 | 0.56 |
ENSMUST00000159109.2
|
Zfp609
|
zinc finger protein 609 |
| chr12_-_21467437 | 0.56 |
ENSMUST00000103002.8
ENSMUST00000155480.9 ENSMUST00000135088.9 |
Ywhaq
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein theta |
| chr5_-_38316296 | 0.56 |
ENSMUST00000201415.4
|
Nsg1
|
neuron specific gene family member 1 |
| chr15_-_103242697 | 0.55 |
ENSMUST00000229373.2
|
Zfp385a
|
zinc finger protein 385A |
| chr19_-_47907705 | 0.55 |
ENSMUST00000095998.7
|
Itprip
|
inositol 1,4,5-triphosphate receptor interacting protein |
| chr6_+_83114086 | 0.55 |
ENSMUST00000087938.11
|
Rtkn
|
rhotekin |
| chr11_-_76514334 | 0.54 |
ENSMUST00000238684.2
|
Abr
|
active BCR-related gene |
| chr3_-_116762476 | 0.54 |
ENSMUST00000119557.8
|
Palmd
|
palmdelphin |
| chr19_-_46315543 | 0.54 |
ENSMUST00000223917.2
ENSMUST00000224447.2 ENSMUST00000041391.5 ENSMUST00000096029.12 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr7_+_126359869 | 0.53 |
ENSMUST00000206272.2
|
Mapk3
|
mitogen-activated protein kinase 3 |
| chr2_-_35322581 | 0.53 |
ENSMUST00000079424.11
|
Ggta1
|
glycoprotein galactosyltransferase alpha 1, 3 |
| chr11_+_67090878 | 0.53 |
ENSMUST00000124516.8
ENSMUST00000018637.15 ENSMUST00000129018.8 |
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr8_+_79755194 | 0.53 |
ENSMUST00000119254.8
ENSMUST00000238669.2 |
Zfp827
|
zinc finger protein 827 |
| chr19_-_5779648 | 0.52 |
ENSMUST00000116558.3
ENSMUST00000099955.4 ENSMUST00000161368.2 |
Fam89b
|
family with sequence similarity 89, member B |
| chr11_+_121128042 | 0.52 |
ENSMUST00000103015.4
|
Narf
|
nuclear prelamin A recognition factor |
| chr17_-_46513499 | 0.52 |
ENSMUST00000024749.9
|
Polh
|
polymerase (DNA directed), eta (RAD 30 related) |
| chr19_+_10019023 | 0.51 |
ENSMUST00000237672.2
|
Fads3
|
fatty acid desaturase 3 |
| chr5_-_134975773 | 0.51 |
ENSMUST00000051401.4
|
Cldn4
|
claudin 4 |
| chr17_+_8144822 | 0.51 |
ENSMUST00000036370.8
|
Tagap
|
T cell activation Rho GTPase activating protein |
| chr10_+_96453408 | 0.51 |
ENSMUST00000218953.2
|
Btg1
|
BTG anti-proliferation factor 1 |
| chr16_+_17269845 | 0.50 |
ENSMUST00000006293.5
ENSMUST00000231228.2 |
Crkl
|
v-crk avian sarcoma virus CT10 oncogene homolog-like |
| chr17_+_36176948 | 0.50 |
ENSMUST00000122899.8
|
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr2_+_91560472 | 0.49 |
ENSMUST00000099712.10
ENSMUST00000111317.9 ENSMUST00000111316.9 ENSMUST00000045705.14 |
Ambra1
|
autophagy/beclin 1 regulator 1 |
| chr7_+_126359763 | 0.49 |
ENSMUST00000091328.4
|
Mapk3
|
mitogen-activated protein kinase 3 |
| chr1_+_39940189 | 0.47 |
ENSMUST00000191761.6
ENSMUST00000193682.6 ENSMUST00000195860.6 ENSMUST00000195259.6 ENSMUST00000195636.6 ENSMUST00000192509.6 |
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr2_+_124452493 | 0.47 |
ENSMUST00000103239.10
ENSMUST00000103240.9 |
Sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr1_-_153363354 | 0.47 |
ENSMUST00000186380.7
ENSMUST00000188345.2 ENSMUST00000042141.12 |
Dhx9
|
DEAH (Asp-Glu-Ala-His) box polypeptide 9 |
| chr3_+_32763313 | 0.47 |
ENSMUST00000126144.3
|
Actl6a
|
actin-like 6A |
| chr3_+_20011405 | 0.47 |
ENSMUST00000108325.9
|
Cp
|
ceruloplasmin |
| chr17_+_34823236 | 0.46 |
ENSMUST00000174041.8
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) |
| chr4_+_62501737 | 0.46 |
ENSMUST00000098031.10
|
Rgs3
|
regulator of G-protein signaling 3 |
| chr15_+_54274151 | 0.45 |
ENSMUST00000036737.4
|
Colec10
|
collectin sub-family member 10 |
| chr15_-_54141816 | 0.45 |
ENSMUST00000079772.4
|
Tnfrsf11b
|
tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) |
| chr12_-_114487525 | 0.44 |
ENSMUST00000103495.3
|
Ighv10-3
|
immunoglobulin heavy variable V10-3 |
| chr10_-_61814852 | 0.43 |
ENSMUST00000105453.8
ENSMUST00000105452.9 ENSMUST00000105454.3 |
Col13a1
|
collagen, type XIII, alpha 1 |
| chr10_-_13264574 | 0.43 |
ENSMUST00000079698.7
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr12_+_24881582 | 0.42 |
ENSMUST00000221952.2
ENSMUST00000078902.8 ENSMUST00000110942.11 |
Mboat2
|
membrane bound O-acyltransferase domain containing 2 |
| chr3_-_143910926 | 0.41 |
ENSMUST00000120539.8
ENSMUST00000196264.5 |
Lmo4
|
LIM domain only 4 |
| chr2_+_4564553 | 0.41 |
ENSMUST00000176828.8
|
Frmd4a
|
FERM domain containing 4A |
| chr17_+_28910393 | 0.40 |
ENSMUST00000124886.9
ENSMUST00000114758.9 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chr15_-_79212323 | 0.40 |
ENSMUST00000166977.9
|
Pla2g6
|
phospholipase A2, group VI |
| chr14_+_34097474 | 0.39 |
ENSMUST00000227130.2
|
Mmrn2
|
multimerin 2 |
| chrX_-_56438380 | 0.38 |
ENSMUST00000143310.2
ENSMUST00000098470.9 ENSMUST00000114726.8 |
Rbmx
|
RNA binding motif protein, X chromosome |
| chr4_+_107224954 | 0.37 |
ENSMUST00000139560.8
|
Ndc1
|
NDC1 transmembrane nucleoporin |
| chr15_-_79212400 | 0.37 |
ENSMUST00000173163.8
ENSMUST00000047816.15 ENSMUST00000172403.9 ENSMUST00000173632.8 |
Pla2g6
|
phospholipase A2, group VI |
| chr14_-_52258158 | 0.37 |
ENSMUST00000228580.2
ENSMUST00000226554.2 ENSMUST00000067549.15 |
Zfp219
|
zinc finger protein 219 |
| chr7_+_65759198 | 0.37 |
ENSMUST00000036372.8
|
Chsy1
|
chondroitin sulfate synthase 1 |
| chr17_-_71158184 | 0.37 |
ENSMUST00000059775.15
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr3_+_20011251 | 0.36 |
ENSMUST00000108328.8
|
Cp
|
ceruloplasmin |
| chr9_-_66032134 | 0.36 |
ENSMUST00000034946.15
|
Snx1
|
sorting nexin 1 |
| chr3_+_32762656 | 0.36 |
ENSMUST00000029214.14
|
Actl6a
|
actin-like 6A |
| chr9_-_57552392 | 0.36 |
ENSMUST00000216934.2
|
Csk
|
c-src tyrosine kinase |
| chr11_-_86964881 | 0.34 |
ENSMUST00000020804.8
|
Gdpd1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr2_+_35472841 | 0.34 |
ENSMUST00000135741.8
|
Dab2ip
|
disabled 2 interacting protein |
| chr2_+_121188195 | 0.34 |
ENSMUST00000125812.8
ENSMUST00000078222.9 ENSMUST00000125221.3 ENSMUST00000150271.8 |
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous |
| chr7_+_88079534 | 0.34 |
ENSMUST00000208478.2
|
Rab38
|
RAB38, member RAS oncogene family |
| chr8_+_108162985 | 0.34 |
ENSMUST00000166615.3
ENSMUST00000213097.2 ENSMUST00000212205.2 |
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr17_-_57394718 | 0.34 |
ENSMUST00000071135.6
|
Tubb4a
|
tubulin, beta 4A class IVA |
| chr3_-_95014090 | 0.34 |
ENSMUST00000005768.8
ENSMUST00000107232.9 ENSMUST00000107236.9 |
Pip5k1a
|
phosphatidylinositol-4-phosphate 5-kinase, type 1 alpha |
| chr18_+_82932747 | 0.33 |
ENSMUST00000071233.7
|
Zfp516
|
zinc finger protein 516 |
| chr3_+_53371086 | 0.33 |
ENSMUST00000058577.5
|
Proser1
|
proline and serine rich 1 |
| chr11_+_114742331 | 0.31 |
ENSMUST00000177952.8
|
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr7_-_44518830 | 0.31 |
ENSMUST00000208682.2
|
Ptov1
|
prostate tumor over expressed gene 1 |
| chr5_-_5315968 | 0.31 |
ENSMUST00000115451.8
ENSMUST00000115452.8 ENSMUST00000131392.8 |
Cdk14
|
cyclin-dependent kinase 14 |
| chr2_+_26205525 | 0.31 |
ENSMUST00000066936.9
ENSMUST00000078616.12 |
Gpsm1
|
G-protein signalling modulator 1 (AGS3-like, C. elegans) |
| chr3_-_116762617 | 0.30 |
ENSMUST00000143611.2
ENSMUST00000040097.14 |
Palmd
|
palmdelphin |
| chr12_+_85043262 | 0.29 |
ENSMUST00000101202.10
|
Ylpm1
|
YLP motif containing 1 |
| chr2_-_20973337 | 0.29 |
ENSMUST00000141298.9
ENSMUST00000125783.3 |
Arhgap21
|
Rho GTPase activating protein 21 |
| chr6_+_88879233 | 0.29 |
ENSMUST00000055022.15
ENSMUST00000204765.3 ENSMUST00000153874.8 |
Tpra1
|
transmembrane protein, adipocyte asscociated 1 |
| chr11_-_74480870 | 0.28 |
ENSMUST00000145524.2
ENSMUST00000102521.9 |
Rap1gap2
|
RAP1 GTPase activating protein 2 |
| chrX_+_162923474 | 0.28 |
ENSMUST00000073973.11
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr1_-_181847492 | 0.28 |
ENSMUST00000177811.8
ENSMUST00000111025.8 ENSMUST00000111024.10 |
Enah
|
ENAH actin regulator |
| chr1_-_4479445 | 0.27 |
ENSMUST00000208660.2
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr2_+_164664920 | 0.27 |
ENSMUST00000132282.2
|
Zswim1
|
zinc finger SWIM-type containing 1 |
| chr8_+_79754980 | 0.27 |
ENSMUST00000087927.11
ENSMUST00000098614.9 |
Zfp827
|
zinc finger protein 827 |
| chr9_+_35334878 | 0.27 |
ENSMUST00000154652.8
|
Cdon
|
cell adhesion molecule-related/down-regulated by oncogenes |
| chr14_+_34097422 | 0.27 |
ENSMUST00000111908.3
|
Mmrn2
|
multimerin 2 |
| chr10_-_22607136 | 0.27 |
ENSMUST00000238910.2
ENSMUST00000127698.8 |
ENSMUSG00000118528.2
Tbpl1
|
novel protein TATA box binding protein-like 1 |
| chr12_-_112640626 | 0.26 |
ENSMUST00000001780.10
|
Akt1
|
thymoma viral proto-oncogene 1 |
| chr11_-_49603501 | 0.26 |
ENSMUST00000020624.7
ENSMUST00000145353.8 |
Cnot6
|
CCR4-NOT transcription complex, subunit 6 |
| chr15_+_76131020 | 0.26 |
ENSMUST00000229380.2
|
Grina
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr10_+_38841511 | 0.26 |
ENSMUST00000019992.6
|
Lama4
|
laminin, alpha 4 |
| chr7_-_26895561 | 0.25 |
ENSMUST00000122202.8
ENSMUST00000080356.10 |
Snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr5_+_34140877 | 0.24 |
ENSMUST00000114382.2
|
Gm1673
|
predicted gene 1673 |
| chr18_-_36330367 | 0.23 |
ENSMUST00000115713.9
|
Nrg2
|
neuregulin 2 |
| chr5_-_33812830 | 0.23 |
ENSMUST00000200849.2
|
Tmem129
|
transmembrane protein 129 |
| chr7_-_26895141 | 0.23 |
ENSMUST00000163311.9
ENSMUST00000126211.2 |
Snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chrX_-_56438322 | 0.23 |
ENSMUST00000114730.8
|
Rbmx
|
RNA binding motif protein, X chromosome |
| chr17_+_37504783 | 0.23 |
ENSMUST00000038844.7
|
Ubd
|
ubiquitin D |
| chr3_+_20011201 | 0.23 |
ENSMUST00000091309.12
ENSMUST00000108329.8 ENSMUST00000003714.13 |
Cp
|
ceruloplasmin |
| chr2_+_138120401 | 0.23 |
ENSMUST00000075410.5
|
Btbd3
|
BTB (POZ) domain containing 3 |
| chr15_+_81350538 | 0.23 |
ENSMUST00000230219.2
|
Rbx1
|
ring-box 1 |
| chr17_-_7228555 | 0.22 |
ENSMUST00000063683.8
|
Tagap1
|
T cell activation GTPase activating protein 1 |
| chr3_-_53370621 | 0.21 |
ENSMUST00000056749.14
|
Nhlrc3
|
NHL repeat containing 3 |
| chr7_-_117728815 | 0.21 |
ENSMUST00000032888.10
|
Arl6ip1
|
ADP-ribosylation factor-like 6 interacting protein 1 |
| chr8_-_61407760 | 0.20 |
ENSMUST00000110302.8
|
Clcn3
|
chloride channel, voltage-sensitive 3 |
| chr6_-_138399896 | 0.20 |
ENSMUST00000161450.8
ENSMUST00000163024.8 ENSMUST00000162185.8 |
Lmo3
|
LIM domain only 3 |
| chr11_+_117223161 | 0.20 |
ENSMUST00000106349.2
|
Septin9
|
septin 9 |
| chr11_+_114743044 | 0.20 |
ENSMUST00000142262.2
|
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr4_+_141473983 | 0.19 |
ENSMUST00000038161.5
|
Agmat
|
agmatine ureohydrolase (agmatinase) |
| chr13_-_6686686 | 0.19 |
ENSMUST00000136585.2
|
Pfkp
|
phosphofructokinase, platelet |
| chr1_+_131080912 | 0.19 |
ENSMUST00000112446.9
ENSMUST00000068805.14 ENSMUST00000068791.11 |
Eif2d
|
eukaryotic translation initiation factor 2D |
| chr3_+_88439616 | 0.19 |
ENSMUST00000172699.2
|
Mex3a
|
mex3 RNA binding family member A |
| chr10_-_4338032 | 0.18 |
ENSMUST00000100078.10
|
Zbtb2
|
zinc finger and BTB domain containing 2 |
| chr4_-_43483696 | 0.18 |
ENSMUST00000030180.7
|
Sit1
|
suppression inducing transmembrane adaptor 1 |
| chr2_+_180240015 | 0.17 |
ENSMUST00000103059.2
|
Col9a3
|
collagen, type IX, alpha 3 |
| chrX_-_48877080 | 0.17 |
ENSMUST00000114893.8
|
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr4_+_57782150 | 0.17 |
ENSMUST00000124581.2
|
Pakap
|
paralemmin A kinase anchor protein |
| chr4_-_126362372 | 0.17 |
ENSMUST00000097888.10
ENSMUST00000239399.2 |
Ago1
|
argonaute RISC catalytic subunit 1 |
| chr11_+_114742619 | 0.16 |
ENSMUST00000053361.12
ENSMUST00000021071.14 ENSMUST00000136785.2 |
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.8 | 2.3 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 0.7 | 2.2 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.6 | 1.9 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.6 | 5.0 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.4 | 1.3 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.3 | 1.4 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.3 | 0.9 | GO:0007225 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.3 | 5.6 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.3 | 1.2 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.3 | 2.2 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.3 | 1.0 | GO:2000657 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.2 | 0.7 | GO:0021693 | cerebellar Purkinje cell layer structural organization(GO:0021693) |
| 0.2 | 0.8 | GO:0003290 | atrial septum secundum morphogenesis(GO:0003290) |
| 0.2 | 0.6 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.2 | 1.3 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.2 | 0.9 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.2 | 1.7 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.2 | 0.6 | GO:1902164 | positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.2 | 0.5 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 0.6 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.1 | 1.1 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.1 | 2.7 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.5 | GO:0042125 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.1 | 0.8 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 1.2 | GO:0033153 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.1 | 0.7 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.1 | 0.6 | GO:1902659 | DNA methylation on cytosine within a CG sequence(GO:0010424) regulation of glucose mediated signaling pathway(GO:1902659) positive regulation of glucose mediated signaling pathway(GO:1902661) |
| 0.1 | 2.9 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 0.7 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.1 | 1.4 | GO:0031659 | positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.1 | 0.5 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 2.6 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 1.3 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 0.5 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.1 | 1.2 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 0.3 | GO:0015827 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) positive regulation of gap junction assembly(GO:1903598) |
| 0.1 | 0.3 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.1 | 0.5 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.1 | 0.5 | GO:0090005 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.1 | 0.2 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 | 0.7 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 0.3 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.8 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 1.0 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 0.5 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 0.3 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.1 | 0.6 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.4 | GO:2001184 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) stress-induced premature senescence(GO:0090400) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.5 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 1.2 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.3 | GO:0090383 | platelet dense granule organization(GO:0060155) phagosome acidification(GO:0090383) melanosome assembly(GO:1903232) |
| 0.0 | 0.5 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.9 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.6 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.7 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.5 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.2 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.4 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.3 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 1.8 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.4 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.6 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 1.1 | GO:0046688 | copper ion transport(GO:0006825) response to copper ion(GO:0046688) |
| 0.0 | 0.5 | GO:0098780 | response to mitochondrial depolarisation(GO:0098780) |
| 0.0 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.3 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.5 | GO:0070571 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 1.7 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 1.6 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 0.2 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.0 | 0.2 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.3 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 2.3 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 2.2 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.4 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.2 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.5 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.5 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.0 | 0.5 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 1.1 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.1 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.6 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 0.3 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.4 | GO:0001942 | hair follicle development(GO:0001942) skin epidermis development(GO:0098773) |
| 0.0 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.9 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.0 | 0.3 | GO:0034308 | primary alcohol metabolic process(GO:0034308) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.5 | 1.9 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 0.3 | 1.3 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.3 | 2.6 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.2 | 0.6 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 1.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.9 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 0.4 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 1.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 1.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.6 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.5 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.1 | 0.3 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.1 | 1.4 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 2.7 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 1.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.3 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 1.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.6 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.0 | 0.6 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.1 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.0 | 0.3 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.8 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 1.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.7 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.1 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 2.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.5 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 1.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.7 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.2 | 0.7 | GO:0034012 | glycerone kinase activity(GO:0004371) FAD-AMP lyase (cyclizing) activity(GO:0034012) triokinase activity(GO:0050354) |
| 0.2 | 1.4 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.2 | 1.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.2 | 0.9 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.2 | 0.7 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.2 | 1.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.2 | 0.5 | GO:0033680 | ATP-dependent DNA/RNA helicase activity(GO:0033680) triplex DNA binding(GO:0045142) |
| 0.1 | 1.2 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.1 | 1.3 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 2.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.4 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.4 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.3 | GO:0036461 | BLOC-2 complex binding(GO:0036461) |
| 0.1 | 1.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.5 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 1.7 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 0.4 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 0.5 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.1 | 0.6 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 1.0 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.8 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 1.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 2.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 1.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 1.0 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.1 | 0.9 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.6 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 1.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 6.1 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.3 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.6 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 1.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.3 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 1.0 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.5 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 2.0 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.6 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) Tat protein binding(GO:0030957) |
| 0.0 | 4.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.3 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.9 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.2 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.4 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 1.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 3.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0102344 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.7 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.8 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 1.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.8 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 1.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.5 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 2.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.3 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 3.4 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 0.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.9 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 1.0 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.0 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 1.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 1.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.4 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.6 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.5 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |