Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Tfap2a
Z-value: 0.74
Transcription factors associated with Tfap2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfap2a
|
ENSMUSG00000021359.17 | Tfap2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfap2a | mm39_v1_chr13_-_40882417_40882421 | 0.18 | 2.9e-01 | Click! |
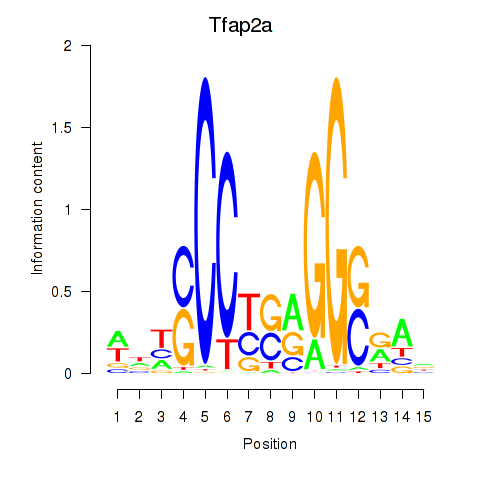
Activity profile of Tfap2a motif
Sorted Z-values of Tfap2a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfap2a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_84387700 | 3.01 |
ENSMUST00000194027.2
ENSMUST00000107689.7 |
Fhdc1
|
FH2 domain containing 1 |
| chr10_+_79716876 | 2.29 |
ENSMUST00000166201.2
|
Prtn3
|
proteinase 3 |
| chr1_-_132294807 | 2.10 |
ENSMUST00000136828.3
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr17_+_48666919 | 2.03 |
ENSMUST00000224001.2
ENSMUST00000024792.8 ENSMUST00000225849.2 |
Treml1
|
triggering receptor expressed on myeloid cells-like 1 |
| chr2_-_131001916 | 1.97 |
ENSMUST00000103188.10
ENSMUST00000133602.8 ENSMUST00000028800.12 |
1700037H04Rik
|
RIKEN cDNA 1700037H04 gene |
| chr19_-_15901919 | 1.79 |
ENSMUST00000162053.8
|
Psat1
|
phosphoserine aminotransferase 1 |
| chr9_+_110687026 | 1.78 |
ENSMUST00000051097.6
|
Prss50
|
protease, serine 50 |
| chr11_+_32246489 | 1.75 |
ENSMUST00000093207.4
|
Hba-a2
|
hemoglobin alpha, adult chain 2 |
| chr19_-_6065181 | 1.74 |
ENSMUST00000236537.2
ENSMUST00000025891.11 |
Capn1
|
calpain 1 |
| chr19_-_6065415 | 1.70 |
ENSMUST00000237519.2
|
Capn1
|
calpain 1 |
| chr11_-_102787950 | 1.69 |
ENSMUST00000067444.10
|
Gfap
|
glial fibrillary acidic protein |
| chr6_+_82923876 | 1.64 |
ENSMUST00000113980.4
|
M1ap
|
meiosis 1 associated protein |
| chr10_+_79715448 | 1.62 |
ENSMUST00000006679.15
|
Prtn3
|
proteinase 3 |
| chr16_+_32427789 | 1.59 |
ENSMUST00000120680.2
|
Tfrc
|
transferrin receptor |
| chr6_-_60805873 | 1.55 |
ENSMUST00000114268.5
|
Snca
|
synuclein, alpha |
| chr9_-_103357564 | 1.53 |
ENSMUST00000124310.5
|
Bfsp2
|
beaded filament structural protein 2, phakinin |
| chr8_+_85428059 | 1.49 |
ENSMUST00000238364.2
ENSMUST00000238562.2 ENSMUST00000037165.6 |
Lyl1
|
lymphoblastomic leukemia 1 |
| chr15_-_103161237 | 1.44 |
ENSMUST00000154510.8
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chrX_-_135116192 | 1.41 |
ENSMUST00000113120.2
ENSMUST00000113118.2 ENSMUST00000058125.9 |
Bex1
|
brain expressed X-linked 1 |
| chr7_-_4815111 | 1.40 |
ENSMUST00000205885.2
|
Ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr9_+_54606144 | 1.34 |
ENSMUST00000120452.8
|
Dnaja4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr10_+_75402090 | 1.33 |
ENSMUST00000129232.8
ENSMUST00000143792.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr12_+_109418759 | 1.32 |
ENSMUST00000056110.15
|
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr16_+_32427738 | 1.31 |
ENSMUST00000023486.15
|
Tfrc
|
transferrin receptor |
| chr3_-_108629594 | 1.25 |
ENSMUST00000029482.16
|
Gpsm2
|
G-protein signalling modulator 2 (AGS3-like, C. elegans) |
| chr7_-_4755971 | 1.21 |
ENSMUST00000183971.8
ENSMUST00000182173.2 ENSMUST00000182738.8 ENSMUST00000182111.8 ENSMUST00000184143.8 ENSMUST00000182048.2 ENSMUST00000063324.14 |
Cox6b2
|
cytochrome c oxidase subunit 6B2 |
| chr8_+_84682136 | 1.20 |
ENSMUST00000005607.9
|
Asf1b
|
anti-silencing function 1B histone chaperone |
| chr2_-_34644376 | 1.15 |
ENSMUST00000142436.2
ENSMUST00000028224.15 ENSMUST00000113099.10 |
Gapvd1
|
GTPase activating protein and VPS9 domains 1 |
| chr11_-_70146156 | 1.12 |
ENSMUST00000108574.3
ENSMUST00000000329.9 |
Alox12
|
arachidonate 12-lipoxygenase |
| chr5_-_24235295 | 1.08 |
ENSMUST00000101513.9
|
Fam126a
|
family with sequence similarity 126, member A |
| chr4_-_140501507 | 1.02 |
ENSMUST00000026381.7
|
Padi4
|
peptidyl arginine deiminase, type IV |
| chr8_+_57964921 | 1.02 |
ENSMUST00000067925.8
|
Hmgb2
|
high mobility group box 2 |
| chr14_-_71003973 | 1.01 |
ENSMUST00000226448.2
ENSMUST00000022696.8 |
Xpo7
|
exportin 7 |
| chr6_+_90596123 | 1.00 |
ENSMUST00000032177.10
|
Slc41a3
|
solute carrier family 41, member 3 |
| chr10_+_11157326 | 1.00 |
ENSMUST00000070300.5
|
Fbxo30
|
F-box protein 30 |
| chr4_-_154110383 | 1.00 |
ENSMUST00000132541.8
ENSMUST00000143047.8 |
Smim1
|
small integral membrane protein 1 |
| chrX_+_92718695 | 0.98 |
ENSMUST00000045898.4
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr11_+_116423266 | 0.98 |
ENSMUST00000106386.8
ENSMUST00000145737.8 ENSMUST00000155102.8 ENSMUST00000063446.13 |
Sphk1
|
sphingosine kinase 1 |
| chr4_-_137157824 | 0.93 |
ENSMUST00000102522.5
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr10_-_5755412 | 0.92 |
ENSMUST00000019907.8
|
Fbxo5
|
F-box protein 5 |
| chr2_-_129139125 | 0.91 |
ENSMUST00000052708.7
|
Ckap2l
|
cytoskeleton associated protein 2-like |
| chr17_-_26160872 | 0.91 |
ENSMUST00000139226.2
ENSMUST00000097368.10 ENSMUST00000140304.2 ENSMUST00000026823.16 |
Pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr17_+_43878989 | 0.90 |
ENSMUST00000167214.8
ENSMUST00000024706.12 |
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr19_+_10019023 | 0.89 |
ENSMUST00000237672.2
|
Fads3
|
fatty acid desaturase 3 |
| chr3_-_137687284 | 0.86 |
ENSMUST00000136613.4
ENSMUST00000029806.13 |
Dapp1
|
dual adaptor for phosphotyrosine and 3-phosphoinositides 1 |
| chr2_-_33777874 | 0.86 |
ENSMUST00000041555.10
|
Mvb12b
|
multivesicular body subunit 12B |
| chr11_-_102787972 | 0.86 |
ENSMUST00000077902.5
|
Gfap
|
glial fibrillary acidic protein |
| chrX_-_7537580 | 0.83 |
ENSMUST00000033486.6
|
Plp2
|
proteolipid protein 2 |
| chr15_-_103123711 | 0.82 |
ENSMUST00000122182.2
ENSMUST00000108813.10 ENSMUST00000127191.2 |
Cbx5
|
chromobox 5 |
| chr13_-_47259652 | 0.81 |
ENSMUST00000021807.13
ENSMUST00000135278.8 |
Dek
|
DEK proto-oncogene (DNA binding) |
| chr7_-_97827461 | 0.80 |
ENSMUST00000040971.14
|
Capn5
|
calpain 5 |
| chr5_-_24235646 | 0.80 |
ENSMUST00000197617.5
ENSMUST00000030849.13 |
Fam126a
|
family with sequence similarity 126, member A |
| chr11_+_79980210 | 0.77 |
ENSMUST00000017694.7
ENSMUST00000108239.7 |
Atad5
|
ATPase family, AAA domain containing 5 |
| chr8_+_57964956 | 0.77 |
ENSMUST00000210871.2
|
Hmgb2
|
high mobility group box 2 |
| chr17_+_71490540 | 0.77 |
ENSMUST00000156570.8
|
Lpin2
|
lipin 2 |
| chr5_-_44139099 | 0.77 |
ENSMUST00000061299.9
|
Fgfbp1
|
fibroblast growth factor binding protein 1 |
| chr16_-_18445172 | 0.77 |
ENSMUST00000231335.2
ENSMUST00000232653.2 |
Gm49601
Septin5
|
predicted gene, 49601 septin 5 |
| chr17_+_43879496 | 0.77 |
ENSMUST00000169694.2
|
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr19_-_9876815 | 0.76 |
ENSMUST00000237147.2
ENSMUST00000025562.9 |
Incenp
|
inner centromere protein |
| chr10_+_11157047 | 0.73 |
ENSMUST00000129456.8
|
Fbxo30
|
F-box protein 30 |
| chr9_-_123768720 | 0.72 |
ENSMUST00000026911.6
|
Ccr1
|
chemokine (C-C motif) receptor 1 |
| chr7_+_127347339 | 0.72 |
ENSMUST00000206893.2
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr14_-_71004019 | 0.72 |
ENSMUST00000167242.8
|
Xpo7
|
exportin 7 |
| chr1_+_91468409 | 0.72 |
ENSMUST00000027538.9
ENSMUST00000190484.7 ENSMUST00000186068.2 |
Asb1
|
ankyrin repeat and SOCS box-containing 1 |
| chr11_+_63022328 | 0.70 |
ENSMUST00000018361.10
|
Pmp22
|
peripheral myelin protein 22 |
| chr10_-_93725619 | 0.69 |
ENSMUST00000181091.8
ENSMUST00000181217.8 ENSMUST00000047910.15 ENSMUST00000180688.2 |
Metap2
|
methionine aminopeptidase 2 |
| chr11_+_50267808 | 0.69 |
ENSMUST00000109142.8
|
Hnrnph1
|
heterogeneous nuclear ribonucleoprotein H1 |
| chr19_+_10582920 | 0.67 |
ENSMUST00000236280.2
|
Ddb1
|
damage specific DNA binding protein 1 |
| chr4_+_149569672 | 0.67 |
ENSMUST00000124413.8
ENSMUST00000141293.8 |
Lzic
|
leucine zipper and CTNNBIP1 domain containing |
| chr19_+_53588808 | 0.66 |
ENSMUST00000025930.10
|
Smc3
|
structural maintenance of chromosomes 3 |
| chr11_-_103844870 | 0.66 |
ENSMUST00000103075.11
|
Nsf
|
N-ethylmaleimide sensitive fusion protein |
| chr7_+_101583283 | 0.65 |
ENSMUST00000209639.2
ENSMUST00000210679.2 |
Numa1
|
nuclear mitotic apparatus protein 1 |
| chr5_-_44139121 | 0.64 |
ENSMUST00000199894.2
|
Fgfbp1
|
fibroblast growth factor binding protein 1 |
| chr19_-_9876745 | 0.64 |
ENSMUST00000237725.2
|
Incenp
|
inner centromere protein |
| chr17_+_27775637 | 0.64 |
ENSMUST00000117254.9
ENSMUST00000231243.2 ENSMUST00000231358.2 ENSMUST00000118570.2 ENSMUST00000231796.2 |
Hmga1
|
high mobility group AT-hook 1 |
| chr12_+_26519203 | 0.62 |
ENSMUST00000020969.5
|
Cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr17_+_27775613 | 0.62 |
ENSMUST00000231780.2
ENSMUST00000232253.2 ENSMUST00000232552.2 ENSMUST00000117600.9 |
Hmga1
|
high mobility group AT-hook 1 |
| chr17_+_27775471 | 0.60 |
ENSMUST00000118599.9
ENSMUST00000232265.2 ENSMUST00000232013.2 ENSMUST00000114888.11 ENSMUST00000231874.2 ENSMUST00000119486.9 ENSMUST00000231825.2 ENSMUST00000231866.2 |
Hmga1
|
high mobility group AT-hook 1 |
| chr8_-_79235505 | 0.60 |
ENSMUST00000211719.2
ENSMUST00000049245.10 |
Rbmxl1
|
RNA binding motif protein, X-linked like-1 |
| chr7_-_126102579 | 0.59 |
ENSMUST00000040202.15
|
Atxn2l
|
ataxin 2-like |
| chr7_-_126102470 | 0.59 |
ENSMUST00000206577.2
|
Atxn2l
|
ataxin 2-like |
| chr5_-_137101108 | 0.59 |
ENSMUST00000077523.4
ENSMUST00000041388.11 |
Serpine1
|
serine (or cysteine) peptidase inhibitor, clade E, member 1 |
| chr6_+_42326714 | 0.59 |
ENSMUST00000203846.3
|
Zyx
|
zyxin |
| chr7_-_3680530 | 0.59 |
ENSMUST00000038743.15
|
Tmc4
|
transmembrane channel-like gene family 4 |
| chr4_+_21776261 | 0.59 |
ENSMUST00000065111.15
ENSMUST00000040429.12 ENSMUST00000148304.8 |
Usp45
|
ubiquitin specific petidase 45 |
| chr2_-_121101822 | 0.59 |
ENSMUST00000110647.8
ENSMUST00000110648.8 |
Trp53bp1
|
transformation related protein 53 binding protein 1 |
| chr17_+_43879366 | 0.59 |
ENSMUST00000167418.8
|
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr9_+_54606832 | 0.58 |
ENSMUST00000070070.8
|
Dnaja4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr19_-_6065799 | 0.58 |
ENSMUST00000235138.2
|
Capn1
|
calpain 1 |
| chr5_+_33815892 | 0.57 |
ENSMUST00000152847.8
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr5_+_108213608 | 0.56 |
ENSMUST00000081567.11
ENSMUST00000170319.8 ENSMUST00000112626.8 |
Mtf2
|
metal response element binding transcription factor 2 |
| chr15_+_9071761 | 0.55 |
ENSMUST00000189437.8
ENSMUST00000190874.8 |
Nadk2
|
NAD kinase 2, mitochondrial |
| chr8_-_111724409 | 0.55 |
ENSMUST00000040416.8
|
Ddx19a
|
DEAD box helicase 19a |
| chr15_+_9071655 | 0.55 |
ENSMUST00000227682.3
|
Nadk2
|
NAD kinase 2, mitochondrial |
| chr17_-_74602469 | 0.55 |
ENSMUST00000233144.2
|
Memo1
|
mediator of cell motility 1 |
| chr17_+_88282472 | 0.54 |
ENSMUST00000005503.5
|
Msh6
|
mutS homolog 6 |
| chr9_+_54606798 | 0.54 |
ENSMUST00000154690.8
|
Dnaja4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr8_-_106578613 | 0.54 |
ENSMUST00000040776.6
|
Cenpt
|
centromere protein T |
| chr2_+_155798457 | 0.54 |
ENSMUST00000109619.9
ENSMUST00000094421.11 ENSMUST00000039994.14 ENSMUST00000151569.8 ENSMUST00000109618.2 |
Cep250
|
centrosomal protein 250 |
| chr8_+_72050292 | 0.54 |
ENSMUST00000143662.8
|
Niban3
|
niban apoptosis regulator 3 |
| chr4_-_43578823 | 0.53 |
ENSMUST00000030189.14
|
Gba2
|
glucosidase beta 2 |
| chr11_-_106679983 | 0.53 |
ENSMUST00000129585.8
|
Ddx5
|
DEAD box helicase 5 |
| chr7_+_127347308 | 0.53 |
ENSMUST00000188580.3
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr14_+_57124028 | 0.53 |
ENSMUST00000223669.2
|
Zmym2
|
zinc finger, MYM-type 2 |
| chr4_+_43578922 | 0.52 |
ENSMUST00000030190.9
|
Rgp1
|
RAB6A GEF compex partner 1 |
| chr1_-_55066629 | 0.52 |
ENSMUST00000027127.14
|
Sf3b1
|
splicing factor 3b, subunit 1 |
| chr2_+_69652714 | 0.51 |
ENSMUST00000053087.4
|
Klhl23
|
kelch-like 23 |
| chr10_+_99851679 | 0.51 |
ENSMUST00000130190.8
ENSMUST00000218200.2 ENSMUST00000020129.8 |
Kitl
|
kit ligand |
| chr5_+_146168020 | 0.51 |
ENSMUST00000161181.8
ENSMUST00000161652.8 ENSMUST00000031640.15 ENSMUST00000159467.2 |
Cdk8
|
cyclin-dependent kinase 8 |
| chr11_+_68936457 | 0.51 |
ENSMUST00000108666.8
ENSMUST00000021277.6 |
Aurkb
|
aurora kinase B |
| chr1_+_172327569 | 0.51 |
ENSMUST00000111230.8
|
Tagln2
|
transgelin 2 |
| chr11_+_50268546 | 0.51 |
ENSMUST00000069304.14
ENSMUST00000077817.8 |
Hnrnph1
|
heterogeneous nuclear ribonucleoprotein H1 |
| chr2_+_69691982 | 0.50 |
ENSMUST00000112260.3
|
Ssb
|
Sjogren syndrome antigen B |
| chr15_-_100322934 | 0.50 |
ENSMUST00000123461.8
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr2_-_156848923 | 0.50 |
ENSMUST00000146413.8
ENSMUST00000103129.9 ENSMUST00000103130.8 |
Dsn1
|
DSN1 homolog, MIS12 kinetochore complex component |
| chr17_-_88105422 | 0.50 |
ENSMUST00000055221.9
|
Kcnk12
|
potassium channel, subfamily K, member 12 |
| chr2_+_4564553 | 0.50 |
ENSMUST00000176828.8
|
Frmd4a
|
FERM domain containing 4A |
| chr11_+_69471219 | 0.50 |
ENSMUST00000108657.4
|
Trp53
|
transformation related protein 53 |
| chr4_+_131600918 | 0.49 |
ENSMUST00000053819.6
|
Srsf4
|
serine and arginine-rich splicing factor 4 |
| chr15_-_96358612 | 0.49 |
ENSMUST00000047835.8
|
Scaf11
|
SR-related CTD-associated factor 11 |
| chr4_+_11191726 | 0.48 |
ENSMUST00000029866.16
ENSMUST00000108324.4 |
Ccne2
|
cyclin E2 |
| chr1_+_172327812 | 0.48 |
ENSMUST00000192460.2
|
Tagln2
|
transgelin 2 |
| chr15_+_78784043 | 0.47 |
ENSMUST00000001226.11
|
Sh3bp1
|
SH3-domain binding protein 1 |
| chr8_+_121463090 | 0.47 |
ENSMUST00000160943.3
ENSMUST00000047737.10 ENSMUST00000162658.8 |
Irf8
|
interferon regulatory factor 8 |
| chr2_-_25498459 | 0.47 |
ENSMUST00000058137.9
|
Rabl6
|
RAB, member RAS oncogene family-like 6 |
| chr2_+_69691906 | 0.46 |
ENSMUST00000090852.11
ENSMUST00000166411.8 |
Ssb
|
Sjogren syndrome antigen B |
| chr8_-_124748124 | 0.46 |
ENSMUST00000165628.9
|
Taf5l
|
TATA-box binding protein associated factor 5 like |
| chr4_-_63965161 | 0.46 |
ENSMUST00000107377.10
|
Tnc
|
tenascin C |
| chr5_+_130200639 | 0.45 |
ENSMUST00000119797.8
ENSMUST00000148264.8 |
Rabgef1
|
RAB guanine nucleotide exchange factor (GEF) 1 |
| chr1_+_180553569 | 0.45 |
ENSMUST00000027780.6
|
Acbd3
|
acyl-Coenzyme A binding domain containing 3 |
| chr7_-_116042674 | 0.44 |
ENSMUST00000170430.3
ENSMUST00000206219.2 |
Pik3c2a
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 alpha |
| chr7_-_130924021 | 0.44 |
ENSMUST00000046611.9
|
Cuzd1
|
CUB and zona pellucida-like domains 1 |
| chr17_-_32569480 | 0.44 |
ENSMUST00000235265.2
ENSMUST00000236503.2 ENSMUST00000050214.9 |
Akap8l
|
A kinase (PRKA) anchor protein 8-like |
| chr4_-_149783097 | 0.44 |
ENSMUST00000038859.14
ENSMUST00000105690.9 |
Pik3cd
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta |
| chr8_+_106412905 | 0.44 |
ENSMUST00000213019.2
|
Carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr11_-_106679671 | 0.44 |
ENSMUST00000123339.2
|
Ddx5
|
DEAD box helicase 5 |
| chr4_-_149783009 | 0.44 |
ENSMUST00000134534.8
ENSMUST00000146612.8 ENSMUST00000105688.10 |
Pik3cd
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta |
| chr14_-_99337137 | 0.44 |
ENSMUST00000042471.11
|
Dis3
|
DIS3 homolog, exosome endoribonuclease and 3'-5' exoribonuclease |
| chr15_-_31601652 | 0.43 |
ENSMUST00000161266.2
|
Cct5
|
chaperonin containing Tcp1, subunit 5 (epsilon) |
| chr8_+_85428391 | 0.43 |
ENSMUST00000238338.2
|
Lyl1
|
lymphoblastomic leukemia 1 |
| chr9_-_66951114 | 0.43 |
ENSMUST00000113686.8
|
Tpm1
|
tropomyosin 1, alpha |
| chr3_+_109030419 | 0.43 |
ENSMUST00000029477.11
|
Slc25a24
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 24 |
| chr7_-_131012202 | 0.43 |
ENSMUST00000207243.2
ENSMUST00000128432.3 ENSMUST00000121033.8 ENSMUST00000046306.15 |
Ikzf5
|
IKAROS family zinc finger 5 |
| chr8_-_71219299 | 0.43 |
ENSMUST00000222087.4
|
Ifi30
|
interferon gamma inducible protein 30 |
| chr9_-_66950991 | 0.43 |
ENSMUST00000113689.8
ENSMUST00000113684.8 |
Tpm1
|
tropomyosin 1, alpha |
| chr16_+_49675682 | 0.43 |
ENSMUST00000114496.3
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr4_-_45320579 | 0.42 |
ENSMUST00000030003.10
|
Exosc3
|
exosome component 3 |
| chr8_+_120729459 | 0.42 |
ENSMUST00000108972.4
|
Crispld2
|
cysteine-rich secretory protein LCCL domain containing 2 |
| chr8_+_69246581 | 0.42 |
ENSMUST00000034328.13
ENSMUST00000110241.8 ENSMUST00000110242.8 ENSMUST00000070713.8 |
Ints10
|
integrator complex subunit 10 |
| chr13_-_95661726 | 0.42 |
ENSMUST00000022185.10
|
F2rl1
|
coagulation factor II (thrombin) receptor-like 1 |
| chr11_+_78213791 | 0.42 |
ENSMUST00000017534.15
|
Aldoc
|
aldolase C, fructose-bisphosphate |
| chr12_-_40088024 | 0.42 |
ENSMUST00000101472.4
|
Arl4a
|
ADP-ribosylation factor-like 4A |
| chr8_+_41692755 | 0.41 |
ENSMUST00000210862.2
ENSMUST00000045218.9 ENSMUST00000211247.2 |
Pcm1
|
pericentriolar material 1 |
| chr11_+_62466851 | 0.41 |
ENSMUST00000102643.2
|
Trpv2
|
transient receptor potential cation channel, subfamily V, member 2 |
| chr5_+_64250268 | 0.41 |
ENSMUST00000087324.7
|
Pgm2
|
phosphoglucomutase 2 |
| chr8_-_103512274 | 0.40 |
ENSMUST00000075190.5
|
Cdh11
|
cadherin 11 |
| chr8_-_85414220 | 0.40 |
ENSMUST00000238449.2
ENSMUST00000238687.2 |
Nacc1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr6_+_42326760 | 0.40 |
ENSMUST00000203652.3
ENSMUST00000070635.13 |
Zyx
|
zyxin |
| chr19_+_46385321 | 0.40 |
ENSMUST00000039922.13
ENSMUST00000111867.9 ENSMUST00000120778.8 |
Sufu
|
SUFU negative regulator of hedgehog signaling |
| chr10_-_21036792 | 0.40 |
ENSMUST00000188495.8
|
Myb
|
myeloblastosis oncogene |
| chr8_-_71249630 | 0.40 |
ENSMUST00000166004.3
|
Mast3
|
microtubule associated serine/threonine kinase 3 |
| chr15_-_75781168 | 0.40 |
ENSMUST00000089680.10
ENSMUST00000141268.8 ENSMUST00000023235.13 ENSMUST00000109972.9 ENSMUST00000089681.12 ENSMUST00000109975.10 ENSMUST00000154584.9 |
Eef1d
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr10_+_7543260 | 0.40 |
ENSMUST00000040135.9
|
Nup43
|
nucleoporin 43 |
| chr13_-_113063890 | 0.39 |
ENSMUST00000022281.5
|
Mtrex
|
Mtr4 exosome RNA helicase |
| chr17_+_24026892 | 0.39 |
ENSMUST00000191385.3
|
Srrm2
|
serine/arginine repetitive matrix 2 |
| chr17_-_71782296 | 0.39 |
ENSMUST00000127430.2
|
Smchd1
|
SMC hinge domain containing 1 |
| chr10_-_76181089 | 0.39 |
ENSMUST00000036033.14
ENSMUST00000160048.8 ENSMUST00000105417.10 |
Dip2a
|
disco interacting protein 2 homolog A |
| chr12_-_105651437 | 0.39 |
ENSMUST00000041055.9
|
Atg2b
|
autophagy related 2B |
| chr7_+_67602565 | 0.39 |
ENSMUST00000005671.10
|
Igf1r
|
insulin-like growth factor I receptor |
| chr6_-_82751429 | 0.38 |
ENSMUST00000000642.11
|
Hk2
|
hexokinase 2 |
| chr15_+_85220438 | 0.38 |
ENSMUST00000163242.3
|
Atxn10
|
ataxin 10 |
| chr7_-_16657825 | 0.38 |
ENSMUST00000019514.10
|
Calm3
|
calmodulin 3 |
| chr5_-_113957318 | 0.38 |
ENSMUST00000201194.4
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr2_-_20973337 | 0.37 |
ENSMUST00000141298.9
ENSMUST00000125783.3 |
Arhgap21
|
Rho GTPase activating protein 21 |
| chr19_-_53932581 | 0.37 |
ENSMUST00000236885.2
ENSMUST00000236098.2 ENSMUST00000236370.2 |
Bbip1
|
BBSome interacting protein 1 |
| chr15_-_75781138 | 0.37 |
ENSMUST00000145764.2
ENSMUST00000116440.9 ENSMUST00000151066.8 |
Eef1d
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr7_+_65710086 | 0.37 |
ENSMUST00000153609.8
|
Snrpa1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr5_-_113957362 | 0.37 |
ENSMUST00000202555.2
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr17_+_29899420 | 0.37 |
ENSMUST00000130052.9
|
Cmtr1
|
cap methyltransferase 1 |
| chr11_-_106084334 | 0.37 |
ENSMUST00000007444.14
ENSMUST00000152008.2 ENSMUST00000103072.10 ENSMUST00000106867.2 |
Strada
|
STE20-related kinase adaptor alpha |
| chr16_-_15455141 | 0.37 |
ENSMUST00000023353.4
|
Mcm4
|
minichromosome maintenance complex component 4 |
| chr3_-_132655804 | 0.36 |
ENSMUST00000117164.8
ENSMUST00000093971.5 ENSMUST00000042729.16 |
Npnt
|
nephronectin |
| chr2_-_163760603 | 0.36 |
ENSMUST00000044734.3
|
Rims4
|
regulating synaptic membrane exocytosis 4 |
| chr5_-_123270449 | 0.36 |
ENSMUST00000186469.7
|
Rhof
|
ras homolog family member F (in filopodia) |
| chr12_-_13299136 | 0.35 |
ENSMUST00000221623.2
|
Ddx1
|
DEAD box helicase 1 |
| chr6_+_42326528 | 0.35 |
ENSMUST00000203329.3
|
Zyx
|
zyxin |
| chr13_-_12272962 | 0.35 |
ENSMUST00000099856.6
|
Mtr
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr19_-_8691460 | 0.35 |
ENSMUST00000206560.2
ENSMUST00000205538.2 |
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chrX_+_12937714 | 0.35 |
ENSMUST00000169594.9
ENSMUST00000089302.11 |
Usp9x
|
ubiquitin specific peptidase 9, X chromosome |
| chr6_-_124710030 | 0.34 |
ENSMUST00000173647.2
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr2_-_44817173 | 0.34 |
ENSMUST00000130991.8
|
Gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr10_+_41685931 | 0.34 |
ENSMUST00000099931.11
|
Sesn1
|
sestrin 1 |
| chr14_+_24540745 | 0.34 |
ENSMUST00000112384.10
|
Rps24
|
ribosomal protein S24 |
| chr6_+_71886030 | 0.34 |
ENSMUST00000055296.11
|
Polr1a
|
polymerase (RNA) I polypeptide A |
| chr1_+_133965228 | 0.34 |
ENSMUST00000162779.2
|
Fmod
|
fibromodulin |
| chr15_-_75781387 | 0.33 |
ENSMUST00000123712.8
ENSMUST00000141475.2 ENSMUST00000144614.8 |
Eef1d
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr11_+_45946800 | 0.33 |
ENSMUST00000011400.8
|
Adam19
|
a disintegrin and metallopeptidase domain 19 (meltrin beta) |
| chr18_-_77802145 | 0.33 |
ENSMUST00000237203.2
|
8030462N17Rik
|
RIKEN cDNA 8030462N17 gene |
| chr12_+_33197753 | 0.33 |
ENSMUST00000146040.9
|
Atxn7l1
|
ataxin 7-like 1 |
| chr8_-_31658775 | 0.33 |
ENSMUST00000033983.6
|
Mak16
|
MAK16 homolog |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.3 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.5 | 1.5 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.4 | 2.2 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.4 | 2.6 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.3 | 0.9 | GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) |
| 0.3 | 1.4 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.3 | 0.5 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.3 | 1.3 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.2 | 1.0 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.2 | 1.6 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.2 | 1.1 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.2 | 1.8 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.2 | 1.3 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.2 | 0.8 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.2 | 1.3 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.2 | 3.3 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.2 | 0.6 | GO:0046072 | dTDP biosynthetic process(GO:0006233) dTDP metabolic process(GO:0046072) |
| 0.2 | 3.9 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.2 | 0.6 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.2 | 1.8 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.2 | 0.5 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.2 | 0.8 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.2 | 0.2 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.2 | 1.3 | GO:1901750 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.2 | 0.5 | GO:1904024 | negative regulation of NAD metabolic process(GO:1902689) negative regulation of glucose catabolic process to lactate via pyruvate(GO:1904024) |
| 0.2 | 0.7 | GO:0055048 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.2 | 1.9 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.2 | 0.2 | GO:0061324 | canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) |
| 0.2 | 2.7 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.2 | 0.9 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.1 | 0.7 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.1 | 0.4 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.1 | 0.4 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.1 | 1.4 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 1.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 1.1 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 0.7 | GO:2000721 | pilomotor reflex(GO:0097195) positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.1 | 0.4 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.1 | 1.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.5 | GO:0051316 | attachment of spindle microtubules to kinetochore involved in meiotic chromosome segregation(GO:0051316) |
| 0.1 | 0.5 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.4 | GO:0034963 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.1 | 0.3 | GO:2000769 | positive regulation of NMDA glutamate receptor activity(GO:1904783) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) |
| 0.1 | 0.5 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.1 | 0.6 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 1.4 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.1 | 1.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.3 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.1 | 0.9 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.4 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 1.5 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 0.5 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 0.3 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.1 | 0.5 | GO:0070668 | mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.1 | 0.2 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.1 | 0.5 | GO:0015692 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 | 2.0 | GO:0045830 | positive regulation of isotype switching(GO:0045830) |
| 0.1 | 0.2 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 1.0 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.2 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.1 | 0.5 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.4 | GO:0035795 | negative regulation of mitochondrial membrane permeability(GO:0035795) positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.1 | 0.6 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.1 | 0.2 | GO:0035037 | sperm entry(GO:0035037) |
| 0.1 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.4 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.1 | 0.3 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.1 | 0.4 | GO:0015867 | ATP transport(GO:0015867) |
| 0.1 | 0.2 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.1 | 0.3 | GO:0060356 | leucine import(GO:0060356) |
| 0.1 | 0.6 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.1 | 0.3 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 0.1 | 0.2 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.2 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 | 0.4 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.1 | 0.3 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 1.2 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 0.7 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.1 | 1.9 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.3 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.3 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.1 | 0.2 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) |
| 0.1 | 0.2 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.1 | 0.1 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.1 | 0.4 | GO:0030397 | regulation of mRNA export from nucleus(GO:0010793) membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.1 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.2 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.5 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.2 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 | 0.2 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.1 | 0.3 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.3 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 0.0 | 0.7 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.2 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.0 | 0.2 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.1 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.0 | 0.4 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.8 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 1.3 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.0 | 0.8 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 2.0 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.1 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 2.3 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.0 | 0.8 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.4 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 1.4 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 1.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.6 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.3 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.1 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 1.4 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.5 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.2 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.3 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.1 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 | 2.1 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.4 | GO:1904871 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.4 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.3 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 | 0.1 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 1.0 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.2 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.0 | 0.1 | GO:0031392 | regulation of prostaglandin biosynthetic process(GO:0031392) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) |
| 0.0 | 0.2 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.7 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.0 | 0.4 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.2 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.0 | GO:0061684 | chaperone-mediated autophagy(GO:0061684) |
| 0.0 | 0.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.7 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.4 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.4 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.0 | 0.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0032803 | low-density lipoprotein particle receptor catabolic process(GO:0032802) regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) |
| 0.0 | 0.3 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 1.5 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.0 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.7 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.2 | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 0.0 | 0.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.3 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.3 | GO:2000637 | positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 2.3 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.3 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.2 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.3 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.0 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.5 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) |
| 0.0 | 0.2 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.8 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:1901204 | regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.0 | 1.7 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.2 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.0 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.0 | 0.1 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.0 | 0.1 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.1 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.1 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 1.6 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.4 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:1900113 | regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.4 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.5 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.1 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.2 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.4 | 1.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.3 | 2.2 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.2 | 1.9 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.2 | 2.9 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 0.5 | GO:0034066 | RIC1-RGP1 guanyl-nucleotide exchange factor complex(GO:0034066) |
| 0.2 | 0.5 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.2 | 0.5 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.2 | 1.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.9 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.7 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.1 | 0.7 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.4 | GO:0071920 | cleavage body(GO:0071920) |
| 0.1 | 0.6 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.6 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.4 | GO:0001740 | Barr body(GO:0001740) |
| 0.1 | 1.0 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 2.7 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.3 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 0.1 | 2.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 1.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.8 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.5 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 2.0 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.1 | 0.4 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 0.7 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.8 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 0.5 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) tubular endosome(GO:0097422) |
| 0.1 | 0.9 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 0.4 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 0.6 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.9 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 1.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.5 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.6 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.4 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.3 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.2 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.9 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 1.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.7 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 1.1 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.3 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.2 | GO:0034448 | EGO complex(GO:0034448) |
| 0.0 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 1.1 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 1.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.7 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.5 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 1.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.2 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.4 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.3 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0031932 | TORC2 complex(GO:0031932) TOR complex(GO:0038201) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.0 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.0 | 1.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.4 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 1.4 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.0 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.9 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.4 | 1.8 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.3 | 1.0 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.3 | 0.8 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.3 | 1.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.3 | 1.3 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.2 | 1.0 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 0.7 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.2 | 2.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.2 | 0.6 | GO:0004798 | thymidylate kinase activity(GO:0004798) |
| 0.2 | 1.9 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.2 | 0.5 | GO:0032142 | single guanine insertion binding(GO:0032142) |
| 0.2 | 1.8 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.2 | 0.7 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.2 | 4.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.7 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 1.5 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.1 | 1.7 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.4 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.1 | 1.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 1.0 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.5 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.9 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.1 | 0.4 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.1 | 0.5 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.8 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.3 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 0.5 | GO:0015639 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.4 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.1 | 0.4 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.1 | 1.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.2 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.1 | 0.9 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 1.3 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 0.2 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 0.4 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 1.9 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.2 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.1 | 0.3 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 0.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 1.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.3 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.1 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.2 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.1 | 0.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.5 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.1 | 1.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.2 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.1 | 0.4 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 1.3 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 0.2 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 1.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.4 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.6 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.6 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.3 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.3 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.7 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.0 | 0.3 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.4 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.4 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.4 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 0.4 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.4 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 1.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.4 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.5 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0033797 | selenate reductase activity(GO:0033797) |
| 0.0 | 0.2 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) |
| 0.0 | 0.6 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 4.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 1.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 1.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.9 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.0 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 1.0 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.1 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.4 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 2.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 1.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.1 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.0 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 2.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.8 | GO:0008026 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 0.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.7 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.1 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.0 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.7 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.1 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 2.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 2.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.5 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.9 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 2.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 3.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.9 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.8 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.7 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.9 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.5 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 1.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.4 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.7 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 1.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 1.7 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 2.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 0.9 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 3.0 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.1 | 1.0 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.8 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.9 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.8 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.0 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 3.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 1.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.7 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 1.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.9 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.5 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.3 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.3 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 1.1 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.0 | 0.4 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.1 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 2.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.7 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.1 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.5 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 1.4 | REACTOME TRANSLATION | Genes involved in Translation |