Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Tfap2c
Z-value: 2.25
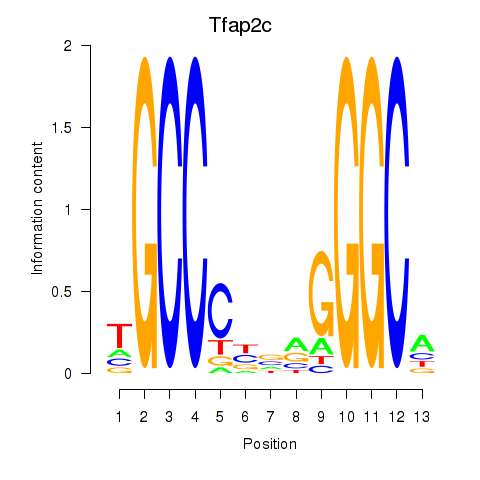
Transcription factors associated with Tfap2c
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfap2c
|
ENSMUSG00000028640.12 | Tfap2c |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfap2c | mm39_v1_chr2_+_172392911_172392968 | 0.29 | 8.6e-02 | Click! |
Activity profile of Tfap2c motif
Sorted Z-values of Tfap2c motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfap2c
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_76756772 | 12.92 |
ENSMUST00000166101.2
|
Sptb
|
spectrin beta, erythrocytic |
| chr14_+_30853010 | 9.29 |
ENSMUST00000227096.2
|
Nt5dc2
|
5'-nucleotidase domain containing 2 |
| chr5_+_33815466 | 9.19 |
ENSMUST00000074849.13
ENSMUST00000079534.11 ENSMUST00000201633.2 |
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr8_+_85628557 | 9.15 |
ENSMUST00000067060.10
ENSMUST00000239392.2 |
Klf1
|
Kruppel-like factor 1 (erythroid) |
| chrX_-_7537580 | 8.74 |
ENSMUST00000033486.6
|
Plp2
|
proteolipid protein 2 |
| chr12_+_109425769 | 7.63 |
ENSMUST00000173812.2
|
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr11_-_102815910 | 7.39 |
ENSMUST00000021311.10
|
Kif18b
|
kinesin family member 18B |
| chr17_+_48666919 | 7.26 |
ENSMUST00000224001.2
ENSMUST00000024792.8 ENSMUST00000225849.2 |
Treml1
|
triggering receptor expressed on myeloid cells-like 1 |
| chr2_+_35146390 | 6.73 |
ENSMUST00000201185.4
ENSMUST00000202990.4 ENSMUST00000202899.4 ENSMUST00000142324.8 ENSMUST00000139867.5 |
Gsn
|
gelsolin |
| chr4_-_116321348 | 6.41 |
ENSMUST00000106486.8
ENSMUST00000106485.8 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr2_+_103788321 | 5.94 |
ENSMUST00000156813.8
ENSMUST00000170926.8 |
Lmo2
|
LIM domain only 2 |
| chr2_+_29759495 | 5.81 |
ENSMUST00000047521.7
ENSMUST00000134152.2 |
Cercam
|
cerebral endothelial cell adhesion molecule |
| chr6_+_4504814 | 5.56 |
ENSMUST00000141483.8
|
Col1a2
|
collagen, type I, alpha 2 |
| chr5_+_137756407 | 5.20 |
ENSMUST00000141733.8
ENSMUST00000110985.2 |
Tsc22d4
|
TSC22 domain family, member 4 |
| chr6_+_4505493 | 4.84 |
ENSMUST00000031668.10
|
Col1a2
|
collagen, type I, alpha 2 |
| chr19_+_5118103 | 4.55 |
ENSMUST00000070630.8
|
Cd248
|
CD248 antigen, endosialin |
| chr10_-_80413119 | 4.49 |
ENSMUST00000038558.9
|
Klf16
|
Kruppel-like factor 16 |
| chr4_+_123176570 | 4.34 |
ENSMUST00000106243.8
ENSMUST00000106241.8 ENSMUST00000080178.13 |
Pabpc4
|
poly(A) binding protein, cytoplasmic 4 |
| chr16_+_32462927 | 4.32 |
ENSMUST00000124585.2
|
Tnk2
|
tyrosine kinase, non-receptor, 2 |
| chr4_+_115696388 | 4.27 |
ENSMUST00000019677.12
ENSMUST00000144427.8 ENSMUST00000106513.10 ENSMUST00000130819.8 ENSMUST00000151203.8 ENSMUST00000140315.2 |
Mknk1
|
MAP kinase-interacting serine/threonine kinase 1 |
| chr11_+_76889415 | 4.27 |
ENSMUST00000108402.9
ENSMUST00000021195.11 |
Slc6a4
|
solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 |
| chr8_-_11058458 | 4.05 |
ENSMUST00000040514.8
|
Irs2
|
insulin receptor substrate 2 |
| chr11_+_3239868 | 4.03 |
ENSMUST00000094471.10
ENSMUST00000110043.8 |
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr17_+_56610321 | 3.91 |
ENSMUST00000001258.15
|
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr17_+_36177498 | 3.84 |
ENSMUST00000187690.7
ENSMUST00000113814.11 |
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr2_-_31973795 | 3.84 |
ENSMUST00000056406.7
|
Fam78a
|
family with sequence similarity 78, member A |
| chr3_-_88410495 | 3.75 |
ENSMUST00000120377.8
ENSMUST00000029699.13 |
Lmna
|
lamin A |
| chr2_-_34803988 | 3.61 |
ENSMUST00000028232.7
ENSMUST00000202907.2 |
Phf19
|
PHD finger protein 19 |
| chr15_+_80507671 | 3.49 |
ENSMUST00000043149.9
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr17_+_56610396 | 3.41 |
ENSMUST00000113038.8
|
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr11_-_75918551 | 3.38 |
ENSMUST00000021207.7
|
Rflnb
|
refilin B |
| chr19_-_3979723 | 3.27 |
ENSMUST00000051803.8
|
Aldh3b1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr8_-_84425724 | 3.20 |
ENSMUST00000005616.16
|
Pkn1
|
protein kinase N1 |
| chr7_-_105436775 | 3.19 |
ENSMUST00000078482.13
|
Dchs1
|
dachsous cadherin related 1 |
| chr2_-_30305313 | 3.12 |
ENSMUST00000132981.9
ENSMUST00000129494.2 |
Crat
|
carnitine acetyltransferase |
| chr2_-_30305472 | 2.96 |
ENSMUST00000134120.2
ENSMUST00000102854.10 |
Crat
|
carnitine acetyltransferase |
| chr15_-_36609208 | 2.96 |
ENSMUST00000001809.15
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr1_-_75455915 | 2.94 |
ENSMUST00000079205.14
ENSMUST00000094818.4 |
Chpf
|
chondroitin polymerizing factor |
| chr4_-_136329953 | 2.91 |
ENSMUST00000105847.8
ENSMUST00000116273.9 |
Kdm1a
|
lysine (K)-specific demethylase 1A |
| chr11_-_69289052 | 2.91 |
ENSMUST00000050140.6
|
Tmem88
|
transmembrane protein 88 |
| chr19_+_7245591 | 2.89 |
ENSMUST00000066646.12
|
Rcor2
|
REST corepressor 2 |
| chr1_-_192883743 | 2.87 |
ENSMUST00000043550.11
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr11_-_78074377 | 2.82 |
ENSMUST00000102483.5
|
Rpl23a
|
ribosomal protein L23A |
| chr11_-_97590460 | 2.81 |
ENSMUST00000103148.8
ENSMUST00000169807.8 |
Pcgf2
|
polycomb group ring finger 2 |
| chr7_+_3339077 | 2.74 |
ENSMUST00000203566.3
|
Myadm
|
myeloid-associated differentiation marker |
| chr8_+_123008855 | 2.70 |
ENSMUST00000054052.15
|
Zfpm1
|
zinc finger protein, multitype 1 |
| chr4_+_140714184 | 2.69 |
ENSMUST00000168047.8
ENSMUST00000037055.14 ENSMUST00000127833.3 |
Atp13a2
|
ATPase type 13A2 |
| chr11_-_59855762 | 2.64 |
ENSMUST00000062405.8
|
Rasd1
|
RAS, dexamethasone-induced 1 |
| chr11_+_77384234 | 2.56 |
ENSMUST00000037285.10
ENSMUST00000100812.4 |
Git1
|
GIT ArfGAP 1 |
| chr8_+_106245368 | 2.48 |
ENSMUST00000034363.7
|
Hsd11b2
|
hydroxysteroid 11-beta dehydrogenase 2 |
| chr9_+_107879700 | 2.48 |
ENSMUST00000035214.11
ENSMUST00000176854.7 ENSMUST00000175874.2 |
Ip6k1
|
inositol hexaphosphate kinase 1 |
| chr2_+_152578164 | 2.47 |
ENSMUST00000038368.9
ENSMUST00000109824.2 |
Id1
|
inhibitor of DNA binding 1, HLH protein |
| chrX_+_55493325 | 2.46 |
ENSMUST00000079663.7
|
Gm2174
|
predicted gene 2174 |
| chr1_-_192883642 | 2.40 |
ENSMUST00000192020.6
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr5_-_148988110 | 2.34 |
ENSMUST00000110505.8
|
Hmgb1
|
high mobility group box 1 |
| chr7_-_100543891 | 2.34 |
ENSMUST00000209041.2
|
Arhgef17
|
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr7_+_24920840 | 2.34 |
ENSMUST00000055604.6
|
Zfp526
|
zinc finger protein 526 |
| chr8_-_85414528 | 2.33 |
ENSMUST00000001975.6
|
Nacc1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr14_-_70945434 | 2.31 |
ENSMUST00000228346.2
|
Xpo7
|
exportin 7 |
| chr17_+_29879569 | 2.30 |
ENSMUST00000024816.13
ENSMUST00000235031.2 ENSMUST00000234911.2 |
Cmtr1
|
cap methyltransferase 1 |
| chr17_+_36176948 | 2.29 |
ENSMUST00000122899.8
|
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr11_-_103247150 | 2.28 |
ENSMUST00000136491.3
ENSMUST00000107023.3 |
Arhgap27
|
Rho GTPase activating protein 27 |
| chr11_+_61575245 | 2.27 |
ENSMUST00000093019.6
|
Fam83g
|
family with sequence similarity 83, member G |
| chr17_+_29879684 | 2.26 |
ENSMUST00000235014.2
ENSMUST00000130423.4 |
Cmtr1
|
cap methyltransferase 1 |
| chr10_-_117681864 | 2.26 |
ENSMUST00000064667.9
|
Rap1b
|
RAS related protein 1b |
| chr11_-_69304501 | 2.25 |
ENSMUST00000094077.5
|
Kdm6b
|
KDM1 lysine (K)-specific demethylase 6B |
| chr4_-_155306992 | 2.17 |
ENSMUST00000084103.10
ENSMUST00000030917.6 |
Ski
|
ski sarcoma viral oncogene homolog (avian) |
| chrX_+_52609954 | 2.16 |
ENSMUST00000063384.4
|
Rtl8c
|
retrotransposon Gag like 8C |
| chr5_-_148988413 | 2.09 |
ENSMUST00000093196.11
|
Hmgb1
|
high mobility group box 1 |
| chr4_-_129436465 | 2.09 |
ENSMUST00000102597.5
|
Hdac1
|
histone deacetylase 1 |
| chr2_-_30305779 | 2.08 |
ENSMUST00000102855.8
ENSMUST00000028207.13 |
Crat
|
carnitine acetyltransferase |
| chr2_-_170269748 | 2.04 |
ENSMUST00000013667.3
ENSMUST00000109152.9 ENSMUST00000068137.11 |
Bcas1
|
breast carcinoma amplified sequence 1 |
| chrX_-_141089165 | 2.04 |
ENSMUST00000134825.3
|
Kcne1l
|
potassium voltage-gated channel, Isk-related family, member 1-like, pseudogene |
| chr10_+_39245746 | 2.03 |
ENSMUST00000063091.13
ENSMUST00000099967.10 ENSMUST00000126486.8 |
Fyn
|
Fyn proto-oncogene |
| chr7_+_89779564 | 2.02 |
ENSMUST00000208742.2
ENSMUST00000049537.9 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr14_+_30973407 | 2.02 |
ENSMUST00000022458.11
|
Bap1
|
Brca1 associated protein 1 |
| chr4_-_141811532 | 2.02 |
ENSMUST00000036572.4
|
Tmem51
|
transmembrane protein 51 |
| chr3_+_104545974 | 2.00 |
ENSMUST00000046212.2
|
Slc16a1
|
solute carrier family 16 (monocarboxylic acid transporters), member 1 |
| chr2_+_130116344 | 1.99 |
ENSMUST00000103198.11
|
Nop56
|
NOP56 ribonucleoprotein |
| chr7_+_105385864 | 1.94 |
ENSMUST00000163389.9
ENSMUST00000136687.9 |
Ilk
|
integrin linked kinase |
| chr9_+_121589044 | 1.94 |
ENSMUST00000093772.4
|
Zfp651
|
zinc finger protein 651 |
| chr5_-_8472582 | 1.90 |
ENSMUST00000168500.8
ENSMUST00000002368.16 |
Dbf4
|
DBF4 zinc finger |
| chr5_-_35836761 | 1.88 |
ENSMUST00000114233.3
|
Htra3
|
HtrA serine peptidase 3 |
| chr2_+_130116357 | 1.88 |
ENSMUST00000136621.9
ENSMUST00000141872.2 |
Nop56
|
NOP56 ribonucleoprotein |
| chr9_-_42035560 | 1.87 |
ENSMUST00000060989.9
|
Sorl1
|
sortilin-related receptor, LDLR class A repeats-containing |
| chr6_-_52141796 | 1.86 |
ENSMUST00000014848.11
|
Hoxa2
|
homeobox A2 |
| chr6_+_88175312 | 1.85 |
ENSMUST00000203480.2
ENSMUST00000015197.9 |
Gata2
|
GATA binding protein 2 |
| chr19_+_10019023 | 1.85 |
ENSMUST00000237672.2
|
Fads3
|
fatty acid desaturase 3 |
| chr11_+_102080446 | 1.84 |
ENSMUST00000070334.10
|
G6pc3
|
glucose 6 phosphatase, catalytic, 3 |
| chr19_+_7394951 | 1.83 |
ENSMUST00000159348.3
|
2700081O15Rik
|
RIKEN cDNA 2700081O15 gene |
| chr19_-_7318798 | 1.81 |
ENSMUST00000165965.8
ENSMUST00000051711.16 ENSMUST00000169541.8 ENSMUST00000165989.2 |
Mark2
|
MAP/microtubule affinity regulating kinase 2 |
| chr8_-_85414220 | 1.78 |
ENSMUST00000238449.2
ENSMUST00000238687.2 |
Nacc1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr13_+_54519161 | 1.78 |
ENSMUST00000026985.9
|
Cplx2
|
complexin 2 |
| chr5_-_123127346 | 1.78 |
ENSMUST00000118027.8
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr11_+_102080489 | 1.71 |
ENSMUST00000078975.8
|
G6pc3
|
glucose 6 phosphatase, catalytic, 3 |
| chr11_+_77923172 | 1.68 |
ENSMUST00000122342.2
ENSMUST00000092881.4 |
Dhrs13
|
dehydrogenase/reductase (SDR family) member 13 |
| chr4_+_134847949 | 1.64 |
ENSMUST00000056977.14
|
Runx3
|
runt related transcription factor 3 |
| chr11_+_69856222 | 1.63 |
ENSMUST00000018713.13
|
Cldn7
|
claudin 7 |
| chr7_+_16550198 | 1.62 |
ENSMUST00000108495.9
|
Strn4
|
striatin, calmodulin binding protein 4 |
| chr11_-_109363406 | 1.61 |
ENSMUST00000168740.3
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr7_-_19338349 | 1.59 |
ENSMUST00000086041.7
|
Clasrp
|
CLK4-associating serine/arginine rich protein |
| chr7_+_127376550 | 1.59 |
ENSMUST00000126761.8
ENSMUST00000047157.13 |
Setd1a
|
SET domain containing 1A |
| chr9_-_121825028 | 1.57 |
ENSMUST00000216669.2
ENSMUST00000215084.2 ENSMUST00000214533.2 ENSMUST00000217610.2 ENSMUST00000084743.7 |
Pomgnt2
|
protein O-linked mannose beta 1,4-N-acetylglucosaminyltransferase 2 |
| chr2_+_118731860 | 1.55 |
ENSMUST00000036578.7
|
Bahd1
|
bromo adjacent homology domain containing 1 |
| chr2_+_156263002 | 1.53 |
ENSMUST00000125153.10
ENSMUST00000103136.8 ENSMUST00000109577.9 |
Epb41l1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr10_-_13744523 | 1.53 |
ENSMUST00000105534.10
|
Aig1
|
androgen-induced 1 |
| chr7_+_89779421 | 1.52 |
ENSMUST00000207225.2
ENSMUST00000207484.2 ENSMUST00000209068.2 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr5_-_123126550 | 1.51 |
ENSMUST00000086200.11
ENSMUST00000156474.8 |
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr3_-_27764522 | 1.49 |
ENSMUST00000195008.6
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr7_+_24982206 | 1.48 |
ENSMUST00000165239.3
|
Cic
|
capicua transcriptional repressor |
| chr10_+_11157326 | 1.44 |
ENSMUST00000070300.5
|
Fbxo30
|
F-box protein 30 |
| chr5_-_123127148 | 1.43 |
ENSMUST00000046073.16
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr5_-_137529251 | 1.40 |
ENSMUST00000132525.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr5_-_8472696 | 1.38 |
ENSMUST00000171808.8
|
Dbf4
|
DBF4 zinc finger |
| chr12_+_110567873 | 1.38 |
ENSMUST00000018851.14
|
Dync1h1
|
dynein cytoplasmic 1 heavy chain 1 |
| chr13_-_34529157 | 1.36 |
ENSMUST00000040336.12
|
Slc22a23
|
solute carrier family 22, member 23 |
| chr16_+_32477722 | 1.36 |
ENSMUST00000238891.2
|
Tnk2
|
tyrosine kinase, non-receptor, 2 |
| chr4_-_46602192 | 1.36 |
ENSMUST00000107756.4
|
Coro2a
|
coronin, actin binding protein 2A |
| chr2_+_128942900 | 1.36 |
ENSMUST00000103205.11
|
Polr1b
|
polymerase (RNA) I polypeptide B |
| chr2_+_128942919 | 1.36 |
ENSMUST00000028874.8
|
Polr1b
|
polymerase (RNA) I polypeptide B |
| chr4_-_86775602 | 1.35 |
ENSMUST00000102814.5
|
Rps6
|
ribosomal protein S6 |
| chr11_+_77923100 | 1.34 |
ENSMUST00000021187.12
|
Dhrs13
|
dehydrogenase/reductase (SDR family) member 13 |
| chr10_-_45346297 | 1.34 |
ENSMUST00000079390.7
|
Lin28b
|
lin-28 homolog B (C. elegans) |
| chr9_-_45923908 | 1.32 |
ENSMUST00000217514.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr1_-_120192977 | 1.30 |
ENSMUST00000140490.8
ENSMUST00000112640.8 |
Steap3
|
STEAP family member 3 |
| chr10_-_13744676 | 1.29 |
ENSMUST00000019942.6
ENSMUST00000162610.8 |
Aig1
|
androgen-induced 1 |
| chr14_+_121272606 | 1.29 |
ENSMUST00000135010.8
|
Farp1
|
FERM, RhoGEF (Arhgef) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr17_+_5542832 | 1.28 |
ENSMUST00000089185.6
|
Zdhhc14
|
zinc finger, DHHC domain containing 14 |
| chr3_-_27764571 | 1.26 |
ENSMUST00000046157.10
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr4_+_138926577 | 1.24 |
ENSMUST00000145368.8
|
Capzb
|
capping protein (actin filament) muscle Z-line, beta |
| chr11_-_106606076 | 1.23 |
ENSMUST00000080853.11
ENSMUST00000183610.8 ENSMUST00000103069.10 ENSMUST00000106796.9 |
Pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr7_+_127078371 | 1.23 |
ENSMUST00000205432.3
|
Fbrs
|
fibrosin |
| chr13_-_49401617 | 1.23 |
ENSMUST00000119721.2
ENSMUST00000058196.13 |
Susd3
|
sushi domain containing 3 |
| chr11_+_3239165 | 1.22 |
ENSMUST00000134089.8
|
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr8_+_3681127 | 1.20 |
ENSMUST00000159911.8
ENSMUST00000004745.9 |
Stxbp2
|
syntaxin binding protein 2 |
| chr16_-_21766282 | 1.19 |
ENSMUST00000023562.9
|
Tmem41a
|
transmembrane protein 41a |
| chr5_-_137529465 | 1.19 |
ENSMUST00000150063.9
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr18_+_74349189 | 1.16 |
ENSMUST00000025444.8
|
Cxxc1
|
CXXC finger 1 (PHD domain) |
| chr11_+_74721733 | 1.15 |
ENSMUST00000000291.9
|
Mnt
|
max binding protein |
| chr2_-_153371861 | 1.12 |
ENSMUST00000035346.14
|
Nol4l
|
nucleolar protein 4-like |
| chrX_+_71006577 | 1.11 |
ENSMUST00000048790.7
|
Prrg3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr2_+_35512023 | 1.10 |
ENSMUST00000091010.12
|
Dab2ip
|
disabled 2 interacting protein |
| chr5_-_123038329 | 1.09 |
ENSMUST00000031435.14
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr4_-_136563154 | 1.09 |
ENSMUST00000105846.9
ENSMUST00000059287.14 ENSMUST00000105845.9 |
Ephb2
|
Eph receptor B2 |
| chr12_+_111725282 | 1.07 |
ENSMUST00000239017.2
ENSMUST00000084941.12 |
Klc1
|
kinesin light chain 1 |
| chr11_-_76737794 | 1.05 |
ENSMUST00000021201.6
|
Cpd
|
carboxypeptidase D |
| chr18_-_33596890 | 1.04 |
ENSMUST00000237066.2
|
Nrep
|
neuronal regeneration related protein |
| chr7_+_24602337 | 1.04 |
ENSMUST00000117796.8
ENSMUST00000047873.16 ENSMUST00000098683.11 ENSMUST00000206508.2 ENSMUST00000206028.2 |
Arhgef1
|
Rho guanine nucleotide exchange factor (GEF) 1 |
| chr19_+_10018914 | 1.01 |
ENSMUST00000115995.4
|
Fads3
|
fatty acid desaturase 3 |
| chr7_-_126101555 | 1.00 |
ENSMUST00000167759.8
|
Atxn2l
|
ataxin 2-like |
| chr12_+_111725357 | 1.00 |
ENSMUST00000118471.8
ENSMUST00000122300.8 |
Klc1
|
kinesin light chain 1 |
| chr7_+_127376267 | 0.98 |
ENSMUST00000144406.8
|
Setd1a
|
SET domain containing 1A |
| chr7_-_16550656 | 0.98 |
ENSMUST00000061390.9
|
Fkrp
|
fukutin related protein |
| chr4_-_135714465 | 0.96 |
ENSMUST00000105851.9
|
Pithd1
|
PITH (C-terminal proteasome-interacting domain of thioredoxin-like) domain containing 1 |
| chrX_+_133657312 | 0.96 |
ENSMUST00000081834.10
ENSMUST00000086880.11 ENSMUST00000086884.5 |
Armcx3
|
armadillo repeat containing, X-linked 3 |
| chr14_+_121148625 | 0.95 |
ENSMUST00000032898.9
|
Ipo5
|
importin 5 |
| chr5_+_117495337 | 0.95 |
ENSMUST00000031309.16
|
Wsb2
|
WD repeat and SOCS box-containing 2 |
| chr7_+_24981604 | 0.93 |
ENSMUST00000163320.8
ENSMUST00000005578.13 |
Cic
|
capicua transcriptional repressor |
| chr19_-_6964988 | 0.93 |
ENSMUST00000130048.8
ENSMUST00000025914.7 |
Vegfb
|
vascular endothelial growth factor B |
| chr7_+_65759198 | 0.92 |
ENSMUST00000036372.8
|
Chsy1
|
chondroitin sulfate synthase 1 |
| chr11_-_70860778 | 0.90 |
ENSMUST00000108530.2
ENSMUST00000035283.11 ENSMUST00000108531.8 |
Nup88
|
nucleoporin 88 |
| chr18_-_33597060 | 0.89 |
ENSMUST00000168890.2
|
Nrep
|
neuronal regeneration related protein |
| chr4_+_44981389 | 0.89 |
ENSMUST00000045078.13
ENSMUST00000128973.2 ENSMUST00000151148.8 |
Grhpr
|
glyoxylate reductase/hydroxypyruvate reductase |
| chr7_+_89779493 | 0.88 |
ENSMUST00000208730.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr6_-_42670021 | 0.87 |
ENSMUST00000121083.8
|
Tcaf1
|
TRPM8 channel-associated factor 1 |
| chr7_-_109559671 | 0.87 |
ENSMUST00000080437.13
|
Dennd5a
|
DENN/MADD domain containing 5A |
| chr11_-_95956176 | 0.85 |
ENSMUST00000100528.5
|
Ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr11_-_106605772 | 0.84 |
ENSMUST00000124958.3
|
Pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr17_-_57338468 | 0.83 |
ENSMUST00000007814.10
ENSMUST00000233480.2 |
Khsrp
|
KH-type splicing regulatory protein |
| chr10_+_11157047 | 0.83 |
ENSMUST00000129456.8
|
Fbxo30
|
F-box protein 30 |
| chr7_-_109559593 | 0.83 |
ENSMUST00000106722.2
|
Dennd5a
|
DENN/MADD domain containing 5A |
| chr15_-_103231921 | 0.81 |
ENSMUST00000229551.2
|
Zfp385a
|
zinc finger protein 385A |
| chr4_+_111272156 | 0.81 |
ENSMUST00000030274.7
|
Bend5
|
BEN domain containing 5 |
| chr2_+_174169351 | 0.79 |
ENSMUST00000124935.8
|
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr12_+_102249294 | 0.79 |
ENSMUST00000056950.14
|
Rin3
|
Ras and Rab interactor 3 |
| chr14_-_70391260 | 0.79 |
ENSMUST00000035612.7
|
Ccar2
|
cell cycle activator and apoptosis regulator 2 |
| chr17_+_5045178 | 0.78 |
ENSMUST00000092723.11
ENSMUST00000232180.2 ENSMUST00000115797.9 |
Arid1b
|
AT rich interactive domain 1B (SWI-like) |
| chr7_-_24937276 | 0.78 |
ENSMUST00000071739.12
ENSMUST00000108411.2 |
Gsk3a
|
glycogen synthase kinase 3 alpha |
| chr17_+_31276649 | 0.77 |
ENSMUST00000236391.2
ENSMUST00000024829.8 ENSMUST00000236427.2 |
Abcg1
|
ATP binding cassette subfamily G member 1 |
| chr3_-_154302679 | 0.77 |
ENSMUST00000052774.8
ENSMUST00000170461.8 ENSMUST00000122976.2 |
Tyw3
|
tRNA-yW synthesizing protein 3 homolog (S. cerevisiae) |
| chr3_-_107993906 | 0.77 |
ENSMUST00000102638.8
ENSMUST00000102637.8 |
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr17_-_13211075 | 0.75 |
ENSMUST00000159986.8
ENSMUST00000007007.14 |
Wtap
|
Wilms tumour 1-associating protein |
| chr17_+_24692858 | 0.74 |
ENSMUST00000054946.10
ENSMUST00000238986.2 ENSMUST00000164508.2 |
Bricd5
|
BRICHOS domain containing 5 |
| chr2_+_156262957 | 0.73 |
ENSMUST00000109574.8
|
Epb41l1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr17_-_46867083 | 0.73 |
ENSMUST00000015749.7
|
Srf
|
serum response factor |
| chr17_-_25104874 | 0.71 |
ENSMUST00000234597.2
|
Nubp2
|
nucleotide binding protein 2 |
| chr2_+_35512172 | 0.70 |
ENSMUST00000112992.9
|
Dab2ip
|
disabled 2 interacting protein |
| chr18_-_33596792 | 0.70 |
ENSMUST00000051087.16
|
Nrep
|
neuronal regeneration related protein |
| chr16_+_32462665 | 0.69 |
ENSMUST00000145627.8
|
Tnk2
|
tyrosine kinase, non-receptor, 2 |
| chr14_+_121272950 | 0.68 |
ENSMUST00000026635.8
|
Farp1
|
FERM, RhoGEF (Arhgef) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr15_-_89033761 | 0.67 |
ENSMUST00000088823.5
|
Mapk11
|
mitogen-activated protein kinase 11 |
| chr4_+_59035088 | 0.66 |
ENSMUST00000041160.13
|
Gng10
|
guanine nucleotide binding protein (G protein), gamma 10 |
| chr6_-_12749192 | 0.66 |
ENSMUST00000172356.8
|
Thsd7a
|
thrombospondin, type I, domain containing 7A |
| chr17_-_30107544 | 0.65 |
ENSMUST00000171691.9
|
Mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr10_+_128295107 | 0.63 |
ENSMUST00000218228.2
|
Smarcc2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
| chr9_+_107445101 | 0.62 |
ENSMUST00000192887.6
ENSMUST00000195752.6 |
Hyal2
|
hyaluronoglucosaminidase 2 |
| chr14_+_20757615 | 0.62 |
ENSMUST00000022358.9
ENSMUST00000224751.2 |
Zswim8
|
zinc finger SWIM-type containing 8 |
| chr11_-_65053710 | 0.61 |
ENSMUST00000093002.12
ENSMUST00000047463.15 |
Arhgap44
|
Rho GTPase activating protein 44 |
| chr18_-_33596468 | 0.61 |
ENSMUST00000171533.9
|
Nrep
|
neuronal regeneration related protein |
| chr2_-_32243295 | 0.60 |
ENSMUST00000091089.12
ENSMUST00000078352.12 ENSMUST00000113350.8 ENSMUST00000202578.4 ENSMUST00000113365.8 |
Dnm1
|
dynamin 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.7 | GO:0097017 | renal protein absorption(GO:0097017) protein processing in phagocytic vesicle(GO:1900756) positive regulation of keratinocyte apoptotic process(GO:1902174) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 1.5 | 4.4 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 1.5 | 5.8 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 1.2 | 3.5 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 1.1 | 7.5 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 1.1 | 4.3 | GO:0021941 | negative regulation of cerebellar granule cell precursor proliferation(GO:0021941) |
| 1.0 | 6.3 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 1.0 | 4.8 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) |
| 0.9 | 4.6 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.8 | 2.5 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.8 | 3.2 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.8 | 3.1 | GO:0017126 | nucleologenesis(GO:0017126) |
| 0.7 | 11.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.7 | 2.1 | GO:0090673 | endothelial cell-matrix adhesion(GO:0090673) |
| 0.7 | 2.7 | GO:0042097 | interleukin-4 biosynthetic process(GO:0042097) regulation of interleukin-4 biosynthetic process(GO:0045402) transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.7 | 2.7 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.7 | 3.3 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.6 | 3.8 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.5 | 2.0 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.5 | 9.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.5 | 5.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.5 | 1.9 | GO:0021658 | rhombomere 3 morphogenesis(GO:0021658) |
| 0.5 | 1.4 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.5 | 7.3 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.4 | 6.4 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.4 | 5.9 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.4 | 3.0 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.4 | 2.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.4 | 3.6 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.4 | 3.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.4 | 9.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) astral microtubule organization(GO:0030953) |
| 0.4 | 1.1 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.4 | 1.8 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.4 | 3.2 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.4 | 3.6 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.3 | 2.1 | GO:2000676 | fungiform papilla formation(GO:0061198) positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.3 | 2.0 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.3 | 4.1 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.3 | 2.5 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.3 | 12.9 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.3 | 2.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.3 | 2.2 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.3 | 1.4 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.3 | 0.8 | GO:1902164 | positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.3 | 1.3 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.3 | 2.3 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 | 5.0 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.2 | 1.3 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.2 | 2.6 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.2 | 2.5 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.2 | 7.6 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.2 | 0.8 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.2 | 2.5 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 0.7 | GO:1903423 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.2 | 0.9 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.2 | 1.6 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.2 | 1.4 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.2 | 2.0 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.2 | 2.6 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.2 | 0.8 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.2 | 0.6 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.2 | 6.4 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 3.3 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.1 | 1.8 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.1 | 2.8 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.1 | 0.8 | GO:0055099 | glycoprotein transport(GO:0034436) response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 0.6 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.1 | 0.4 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.1 | 0.6 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 | 0.9 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.1 | 7.4 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 3.9 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.1 | 0.3 | GO:0030719 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 0.1 | 2.6 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.1 | 1.0 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.4 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.1 | 0.9 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 1.2 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.1 | 0.7 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 0.4 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) |
| 0.1 | 0.8 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 1.6 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 1.6 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 0.7 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 4.5 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 2.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 1.9 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 0.8 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.2 | GO:0061643 | chemorepulsion of axon(GO:0061643) |
| 0.1 | 2.7 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.1 | 0.8 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.1 | 0.5 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 | 1.1 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.5 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) |
| 0.1 | 4.3 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.1 | 6.7 | GO:0030168 | platelet activation(GO:0030168) |
| 0.1 | 1.0 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 2.5 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 0.8 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.5 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.5 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 1.6 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 1.3 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.4 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 1.9 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 2.4 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 3.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.3 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 1.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.3 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 4.1 | GO:0008584 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) otic vesicle formation(GO:0030916) |
| 0.0 | 1.3 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.5 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.2 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.0 | 2.4 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.2 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.0 | 0.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 1.3 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 1.7 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 1.7 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 1.0 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 2.2 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.0 | 0.7 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.0 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.3 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.0 | 0.5 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.5 | GO:0003170 | heart valve development(GO:0003170) |
| 0.0 | 0.4 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 2.2 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.1 | GO:0030578 | PML body organization(GO:0030578) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.4 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 1.6 | 12.9 | GO:0008091 | spectrin(GO:0008091) |
| 0.7 | 6.4 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.7 | 6.7 | GO:0030478 | actin cap(GO:0030478) |
| 0.7 | 7.4 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.6 | 4.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.5 | 3.8 | GO:0005638 | lamin filament(GO:0005638) |
| 0.4 | 1.8 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.4 | 2.7 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.4 | 3.9 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.4 | 1.8 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.2 | 7.3 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.2 | 3.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 1.2 | GO:0070820 | cytolytic granule(GO:0044194) tertiary granule(GO:0070820) |
| 0.2 | 2.8 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.2 | 2.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.2 | 3.7 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.2 | 2.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 2.8 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.2 | 1.6 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.2 | 10.0 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 1.3 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 0.8 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 1.4 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 0.4 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 7.7 | GO:0000791 | euchromatin(GO:0000791) |
| 0.1 | 2.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 3.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.9 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.8 | GO:0036396 | MIS complex(GO:0036396) |
| 0.1 | 2.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 1.2 | GO:0071203 | F-actin capping protein complex(GO:0008290) WASH complex(GO:0071203) |
| 0.1 | 4.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 4.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 2.9 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 0.5 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 7.8 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 0.5 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 3.2 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 1.7 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 1.0 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 5.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.4 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.4 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.8 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 1.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.9 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 12.8 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 1.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.7 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 5.4 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 9.6 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 1.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 2.1 | GO:0001650 | fibrillar center(GO:0001650) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.5 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 1.7 | 6.7 | GO:0045159 | myosin II binding(GO:0045159) |
| 1.5 | 4.4 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 1.4 | 4.3 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 1.3 | 3.9 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 1.0 | 7.3 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.9 | 3.6 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.8 | 2.5 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.8 | 3.2 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.8 | 4.6 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.7 | 2.1 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.6 | 10.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.6 | 2.9 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.5 | 2.6 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.4 | 2.5 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.4 | 2.0 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.4 | 2.8 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.4 | 1.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.4 | 5.8 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.4 | 9.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.4 | 3.3 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.3 | 2.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.3 | 0.9 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.3 | 4.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.3 | 1.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.3 | 4.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.3 | 11.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.3 | 1.8 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.3 | 8.7 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.2 | 2.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.2 | 2.8 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.2 | 7.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 2.0 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 3.9 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 2.7 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.2 | 0.8 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.2 | 1.3 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.2 | 1.9 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.2 | 2.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 0.2 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.2 | 1.3 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 0.6 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 1.6 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 1.7 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 0.4 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.1 | 2.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 17.8 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 3.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.5 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 6.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 2.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.9 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 2.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.9 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 1.0 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 7.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.7 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.1 | 1.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.7 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.1 | 0.8 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 1.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.3 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 1.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 2.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.8 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 3.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 2.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 3.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 14.4 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 2.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 1.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 3.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 1.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.5 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 1.0 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 2.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.6 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.4 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 4.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.6 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.1 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.6 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 15.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.2 | 3.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 3.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 8.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 2.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 5.1 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 0.9 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 14.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 5.7 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 3.5 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 2.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 2.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 2.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 4.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 3.6 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 5.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.1 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 1.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 3.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 12.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.3 | 4.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 12.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 6.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 7.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.2 | 4.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.2 | 3.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 7.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 2.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 2.3 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 2.0 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 4.8 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 4.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 2.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 3.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 2.7 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 0.9 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 3.3 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.1 | 3.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 3.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.5 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 2.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 0.7 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 2.8 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 0.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 2.8 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 3.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 5.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 1.3 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 2.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.4 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 2.3 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |