Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Tfap2e
Z-value: 0.48
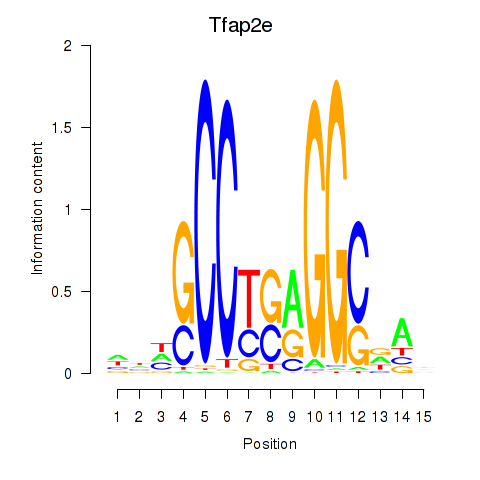
Transcription factors associated with Tfap2e
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfap2e
|
ENSMUSG00000042477.8 | Tfap2e |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfap2e | mm39_v1_chr4_-_126630028_126630039 | 0.20 | 2.4e-01 | Click! |
Activity profile of Tfap2e motif
Sorted Z-values of Tfap2e motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfap2e
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_46146558 | 4.34 |
ENSMUST00000121916.8
ENSMUST00000034586.9 |
Apoc3
|
apolipoprotein C-III |
| chr9_-_46146928 | 3.69 |
ENSMUST00000118649.8
|
Apoc3
|
apolipoprotein C-III |
| chr2_+_102488985 | 1.71 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr4_+_118217179 | 1.52 |
ENSMUST00000006562.6
|
Hyi
|
hydroxypyruvate isomerase (putative) |
| chr5_-_34345014 | 1.30 |
ENSMUST00000042701.13
ENSMUST00000119171.2 |
Mxd4
|
Max dimerization protein 4 |
| chr4_-_57143437 | 0.91 |
ENSMUST00000095076.10
ENSMUST00000030142.4 |
Epb41l4b
|
erythrocyte membrane protein band 4.1 like 4b |
| chr16_-_46317135 | 0.91 |
ENSMUST00000149901.2
ENSMUST00000096052.9 |
Nectin3
|
nectin cell adhesion molecule 3 |
| chr14_+_57124028 | 0.89 |
ENSMUST00000223669.2
|
Zmym2
|
zinc finger, MYM-type 2 |
| chr17_+_64170967 | 0.87 |
ENSMUST00000232945.2
|
Fer
|
fer (fms/fps related) protein kinase |
| chr6_+_108190163 | 0.86 |
ENSMUST00000203615.3
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor 1 |
| chr6_+_54016543 | 0.83 |
ENSMUST00000046856.14
|
Chn2
|
chimerin 2 |
| chr6_+_108190050 | 0.78 |
ENSMUST00000032192.9
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor 1 |
| chr11_-_51647204 | 0.77 |
ENSMUST00000109092.8
ENSMUST00000064297.5 |
Sec24a
|
Sec24 related gene family, member A (S. cerevisiae) |
| chr2_+_133394079 | 0.77 |
ENSMUST00000028836.7
|
Bmp2
|
bone morphogenetic protein 2 |
| chr11_-_51647290 | 0.65 |
ENSMUST00000109097.9
|
Sec24a
|
Sec24 related gene family, member A (S. cerevisiae) |
| chr9_+_100525637 | 0.62 |
ENSMUST00000041418.13
|
Stag1
|
stromal antigen 1 |
| chr9_+_100525807 | 0.61 |
ENSMUST00000133388.2
|
Stag1
|
stromal antigen 1 |
| chr17_-_64170786 | 0.59 |
ENSMUST00000050753.4
|
A930002H24Rik
|
RIKEN cDNA A930002H24 gene |
| chr8_+_108020132 | 0.55 |
ENSMUST00000151114.8
ENSMUST00000125721.8 ENSMUST00000075922.11 |
Nfat5
|
nuclear factor of activated T cells 5 |
| chr9_+_100525501 | 0.54 |
ENSMUST00000146312.8
ENSMUST00000129269.8 |
Stag1
|
stromal antigen 1 |
| chr11_-_109613040 | 0.52 |
ENSMUST00000020938.8
|
Fam20a
|
FAM20A, golgi associated secretory pathway pseudokinase |
| chr8_+_108020092 | 0.49 |
ENSMUST00000169453.8
|
Nfat5
|
nuclear factor of activated T cells 5 |
| chr9_+_109760856 | 0.48 |
ENSMUST00000169851.8
|
Map4
|
microtubule-associated protein 4 |
| chr6_+_56691875 | 0.47 |
ENSMUST00000031805.11
ENSMUST00000177249.3 |
Avl9
|
AVL9 cell migration associated |
| chr9_+_109760931 | 0.46 |
ENSMUST00000165876.8
|
Map4
|
microtubule-associated protein 4 |
| chr7_-_116042674 | 0.42 |
ENSMUST00000170430.3
ENSMUST00000206219.2 |
Pik3c2a
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 alpha |
| chr2_+_83642910 | 0.40 |
ENSMUST00000051454.4
|
Fam171b
|
family with sequence similarity 171, member B |
| chrX_+_12937714 | 0.39 |
ENSMUST00000169594.9
ENSMUST00000089302.11 |
Usp9x
|
ubiquitin specific peptidase 9, X chromosome |
| chr11_+_95304903 | 0.37 |
ENSMUST00000107724.9
ENSMUST00000150884.8 ENSMUST00000107722.8 ENSMUST00000127713.2 |
Spop
|
speckle-type BTB/POZ protein |
| chr13_-_111626562 | 0.37 |
ENSMUST00000091236.11
ENSMUST00000047627.14 |
Gpbp1
|
GC-rich promoter binding protein 1 |
| chrX_+_7588505 | 0.37 |
ENSMUST00000207675.2
ENSMUST00000116633.9 ENSMUST00000208996.2 ENSMUST00000144148.4 ENSMUST00000125991.9 ENSMUST00000148624.8 |
Wdr45
|
WD repeat domain 45 |
| chr2_+_109721189 | 0.35 |
ENSMUST00000028583.8
|
Lin7c
|
lin-7 homolog C (C. elegans) |
| chr8_+_41692755 | 0.34 |
ENSMUST00000210862.2
ENSMUST00000045218.9 ENSMUST00000211247.2 |
Pcm1
|
pericentriolar material 1 |
| chr13_+_38009981 | 0.32 |
ENSMUST00000110238.10
|
Rreb1
|
ras responsive element binding protein 1 |
| chr8_+_80366247 | 0.32 |
ENSMUST00000173078.8
ENSMUST00000173286.8 |
Otud4
|
OTU domain containing 4 |
| chrX_+_7589166 | 0.31 |
ENSMUST00000115687.8
|
Wdr45
|
WD repeat domain 45 |
| chrX_+_72760318 | 0.30 |
ENSMUST00000114461.3
|
Abcd1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr14_+_73380577 | 0.29 |
ENSMUST00000165567.8
ENSMUST00000022702.13 |
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr15_-_38299934 | 0.29 |
ENSMUST00000228772.2
|
Klf10
|
Kruppel-like factor 10 |
| chr6_-_39702381 | 0.29 |
ENSMUST00000002487.15
|
Braf
|
Braf transforming gene |
| chr3_-_54962899 | 0.28 |
ENSMUST00000199144.5
|
Ccna1
|
cyclin A1 |
| chr7_-_30298287 | 0.28 |
ENSMUST00000108150.2
|
Zbtb32
|
zinc finger and BTB domain containing 32 |
| chr13_+_38010203 | 0.28 |
ENSMUST00000128570.9
|
Rreb1
|
ras responsive element binding protein 1 |
| chr3_-_88362606 | 0.27 |
ENSMUST00000125526.8
|
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr16_-_35589726 | 0.27 |
ENSMUST00000023554.9
|
Slc49a4
|
solute carrier family 49 member 4 |
| chr9_+_108394351 | 0.26 |
ENSMUST00000192932.6
ENSMUST00000193348.2 ENSMUST00000194385.2 |
Qrich1
|
glutamine-rich 1 |
| chr5_-_149559792 | 0.26 |
ENSMUST00000202361.4
ENSMUST00000202089.4 ENSMUST00000200825.2 ENSMUST00000201559.4 |
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr7_+_80707328 | 0.25 |
ENSMUST00000107348.2
|
Alpk3
|
alpha-kinase 3 |
| chr3_-_54962922 | 0.25 |
ENSMUST00000197238.5
|
Ccna1
|
cyclin A1 |
| chr14_+_73380539 | 0.24 |
ENSMUST00000169513.8
ENSMUST00000165727.8 |
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr9_+_108394005 | 0.23 |
ENSMUST00000195563.6
|
Qrich1
|
glutamine-rich 1 |
| chrX_+_72760183 | 0.23 |
ENSMUST00000002084.14
|
Abcd1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr5_-_149559737 | 0.22 |
ENSMUST00000200805.4
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr9_+_108394761 | 0.21 |
ENSMUST00000192819.2
|
Qrich1
|
glutamine-rich 1 |
| chrX_-_103024847 | 0.21 |
ENSMUST00000121153.8
ENSMUST00000070705.6 |
Rlim
|
ring finger protein, LIM domain interacting |
| chr1_-_177624017 | 0.19 |
ENSMUST00000016105.9
|
Adss
|
adenylosuccinate synthetase, non muscle |
| chr13_+_38009951 | 0.19 |
ENSMUST00000138043.8
|
Rreb1
|
ras responsive element binding protein 1 |
| chr7_+_101916992 | 0.19 |
ENSMUST00000033289.6
ENSMUST00000209255.2 |
Stim1
|
stromal interaction molecule 1 |
| chr14_-_14371476 | 0.19 |
ENSMUST00000225653.2
ENSMUST00000112689.9 ENSMUST00000036682.9 |
Pxk
|
PX domain containing serine/threonine kinase |
| chr2_-_38177359 | 0.19 |
ENSMUST00000102787.10
|
Dennd1a
|
DENN/MADD domain containing 1A |
| chr4_+_148123554 | 0.18 |
ENSMUST00000141283.8
|
Mthfr
|
methylenetetrahydrofolate reductase |
| chr6_-_39702127 | 0.18 |
ENSMUST00000101497.4
|
Braf
|
Braf transforming gene |
| chr3_-_94693740 | 0.17 |
ENSMUST00000153263.9
ENSMUST00000107272.7 ENSMUST00000155485.4 |
Cgn
|
cingulin |
| chr12_+_111540920 | 0.17 |
ENSMUST00000075281.8
ENSMUST00000084953.13 |
Mark3
|
MAP/microtubule affinity regulating kinase 3 |
| chr6_-_136638926 | 0.17 |
ENSMUST00000032336.7
|
Plbd1
|
phospholipase B domain containing 1 |
| chr12_-_45120895 | 0.16 |
ENSMUST00000120531.8
ENSMUST00000143376.8 |
Stxbp6
|
syntaxin binding protein 6 (amisyn) |
| chr15_+_80862074 | 0.16 |
ENSMUST00000229727.2
|
Sgsm3
|
small G protein signaling modulator 3 |
| chr9_+_21077010 | 0.15 |
ENSMUST00000039413.15
|
Pde4a
|
phosphodiesterase 4A, cAMP specific |
| chr2_-_150746574 | 0.15 |
ENSMUST00000056149.15
|
Abhd12
|
abhydrolase domain containing 12 |
| chr3_+_103187162 | 0.15 |
ENSMUST00000106860.6
|
Trim33
|
tripartite motif-containing 33 |
| chr3_-_113423774 | 0.15 |
ENSMUST00000092154.10
ENSMUST00000106536.8 ENSMUST00000106535.2 |
Rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr2_-_173117936 | 0.15 |
ENSMUST00000139306.2
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr9_+_25163735 | 0.15 |
ENSMUST00000115272.9
ENSMUST00000165594.4 |
Septin7
|
septin 7 |
| chr8_+_94879235 | 0.14 |
ENSMUST00000034211.10
ENSMUST00000211930.2 ENSMUST00000211915.2 |
Mt3
|
metallothionein 3 |
| chr3_+_89680867 | 0.13 |
ENSMUST00000038356.13
|
Ube2q1
|
ubiquitin-conjugating enzyme E2Q family member 1 |
| chr2_+_27055245 | 0.13 |
ENSMUST00000000910.7
|
Dbh
|
dopamine beta hydroxylase |
| chr1_+_180553569 | 0.12 |
ENSMUST00000027780.6
|
Acbd3
|
acyl-Coenzyme A binding domain containing 3 |
| chr5_-_149559636 | 0.12 |
ENSMUST00000201452.4
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr11_-_102109748 | 0.10 |
ENSMUST00000131254.2
|
Hdac5
|
histone deacetylase 5 |
| chr9_-_13245834 | 0.10 |
ENSMUST00000110582.4
|
Jrkl
|
Jrk-like |
| chr5_-_149559667 | 0.10 |
ENSMUST00000074846.14
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr6_+_136804828 | 0.09 |
ENSMUST00000204086.2
|
BC049715
|
cDNA sequence BC049715 |
| chr10_+_85763545 | 0.09 |
ENSMUST00000170396.3
|
Ascl4
|
achaete-scute family bHLH transcription factor 4 |
| chr17_+_37356854 | 0.09 |
ENSMUST00000025338.16
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr1_+_74811045 | 0.09 |
ENSMUST00000006716.8
|
Wnt6
|
wingless-type MMTV integration site family, member 6 |
| chr9_+_108394269 | 0.08 |
ENSMUST00000195513.6
ENSMUST00000193258.6 ENSMUST00000112155.9 ENSMUST00000006851.15 |
Qrich1
|
glutamine-rich 1 |
| chr4_-_127247864 | 0.07 |
ENSMUST00000106090.8
ENSMUST00000060419.2 |
Gjb4
|
gap junction protein, beta 4 |
| chr11_+_49916136 | 0.07 |
ENSMUST00000054684.14
ENSMUST00000238748.2 ENSMUST00000102776.5 |
Rnf130
|
ring finger protein 130 |
| chr11_-_100139728 | 0.06 |
ENSMUST00000007280.9
|
Krt16
|
keratin 16 |
| chr9_-_119812042 | 0.06 |
ENSMUST00000214058.2
|
Csrnp1
|
cysteine-serine-rich nuclear protein 1 |
| chr16_+_93629009 | 0.06 |
ENSMUST00000044068.10
ENSMUST00000202261.5 ENSMUST00000201097.3 |
Morc3
|
microrchidia 3 |
| chr6_-_137626207 | 0.05 |
ENSMUST00000134630.6
ENSMUST00000058210.13 ENSMUST00000111878.8 |
Eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr12_-_114186874 | 0.04 |
ENSMUST00000103477.4
ENSMUST00000192499.3 |
Ighv7-4
|
immunoglobulin heavy variable 7-4 |
| chr11_+_98277276 | 0.03 |
ENSMUST00000041301.8
|
Pnmt
|
phenylethanolamine-N-methyltransferase |
| chr5_-_146521629 | 0.03 |
ENSMUST00000200112.2
ENSMUST00000197431.2 ENSMUST00000197825.2 |
Gpr12
|
G-protein coupled receptor 12 |
| chr2_+_31204314 | 0.03 |
ENSMUST00000113532.9
|
Hmcn2
|
hemicentin 2 |
| chr15_+_89383799 | 0.03 |
ENSMUST00000109309.9
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr12_+_4967376 | 0.03 |
ENSMUST00000045664.7
|
Atad2b
|
ATPase family, AAA domain containing 2B |
| chr3_+_60408678 | 0.02 |
ENSMUST00000191747.6
ENSMUST00000194069.6 |
Mbnl1
|
muscleblind like splicing factor 1 |
| chr7_-_19363280 | 0.02 |
ENSMUST00000094762.10
ENSMUST00000049912.15 ENSMUST00000098754.5 |
Relb
|
avian reticuloendotheliosis viral (v-rel) oncogene related B |
| chr11_-_97635484 | 0.02 |
ENSMUST00000018691.9
|
Pip4k2b
|
phosphatidylinositol-5-phosphate 4-kinase, type II, beta |
| chr7_-_19362328 | 0.01 |
ENSMUST00000141586.2
ENSMUST00000153309.8 |
Relb
|
avian reticuloendotheliosis viral (v-rel) oncogene related B |
| chr7_-_103832599 | 0.01 |
ENSMUST00000216612.3
|
Olfr648
|
olfactory receptor 648 |
| chr6_-_136804728 | 0.01 |
ENSMUST00000146348.4
|
Wbp11
|
WW domain binding protein 11 |
| chr4_+_42158092 | 0.01 |
ENSMUST00000098122.3
|
Gm13306
|
predicted gene 13306 |
| chr7_-_44174065 | 0.00 |
ENSMUST00000165208.4
|
Mybpc2
|
myosin binding protein C, fast-type |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 8.0 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.2 | 1.7 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.2 | 0.8 | GO:0003130 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 0.2 | 1.6 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.4 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.1 | 0.9 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.5 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 0.8 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.9 | GO:0038109 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Fc-epsilon receptor signaling pathway(GO:0038095) Kit signaling pathway(GO:0038109) |
| 0.1 | 1.4 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 0.4 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.1 | 0.9 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.1 | 0.7 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.1 | 0.4 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.5 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.2 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.0 | 0.3 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.7 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.1 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.9 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.1 | GO:0046333 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.0 | 0.2 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.7 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 1.0 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.2 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.0 | 0.0 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0051546 | keratinocyte migration(GO:0051546) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 8.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.4 | 1.6 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.3 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.1 | 1.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.0 | 0.7 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 1.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0000242 | pericentriolar material(GO:0000242) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 8.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.3 | 1.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.3 | 1.7 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.8 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 0.2 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.1 | 0.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 0.5 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.9 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.5 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.8 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.0 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 1.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.5 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 1.7 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |