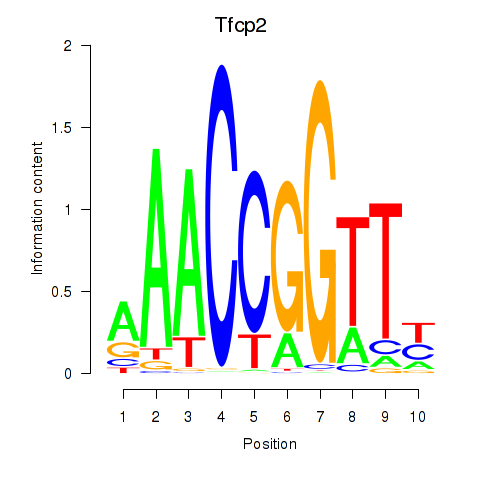
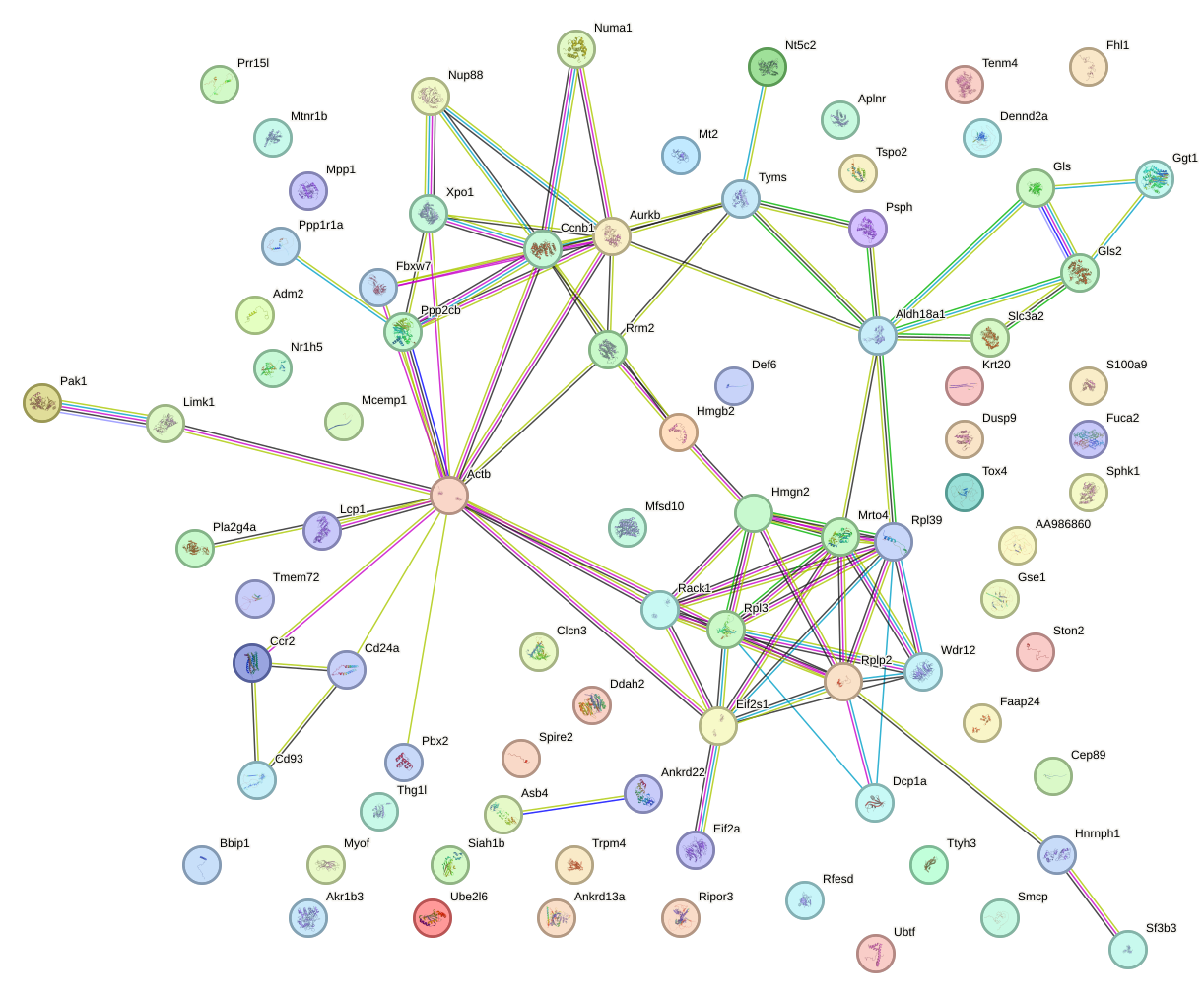
|
chr10_+_43455919
Show fit
|
41.33 |
ENSMUST00000214476.2
|
Cd24a
|
CD24a antigen
|
|
chr8_+_94899292
Show fit
|
34.69 |
ENSMUST00000034214.8
ENSMUST00000212806.2
|
Mt2
|
metallothionein 2
|
|
chr10_+_43455157
Show fit
|
29.44 |
ENSMUST00000058714.10
|
Cd24a
|
CD24a antigen
|
|
chr17_-_48758538
Show fit
|
24.73 |
ENSMUST00000024794.12
|
Tspo2
|
translocator protein 2
|
|
chr3_-_90603013
Show fit
|
23.70 |
ENSMUST00000069960.12
ENSMUST00000117167.2
|
S100a9
|
S100 calcium binding protein A9 (calgranulin B)
|
|
chr8_+_57964921
Show fit
|
21.51 |
ENSMUST00000067925.8
|
Hmgb2
|
high mobility group box 2
|
|
chr8_+_57964956
Show fit
|
18.31 |
ENSMUST00000210871.2
|
Hmgb2
|
high mobility group box 2
|
|
chr11_+_68936457
Show fit
|
17.89 |
ENSMUST00000108666.8
ENSMUST00000021277.6
|
Aurkb
|
aurora kinase B
|
|
chr11_+_116423266
Show fit
|
15.27 |
ENSMUST00000106386.8
ENSMUST00000145737.8
ENSMUST00000155102.8
ENSMUST00000063446.13
|
Sphk1
|
sphingosine kinase 1
|
|
chr13_-_100922910
Show fit
|
14.11 |
ENSMUST00000174038.2
ENSMUST00000091295.14
ENSMUST00000072119.15
|
Ccnb1
|
cyclin B1
|
|
chr10_+_75407356
Show fit
|
13.33 |
ENSMUST00000143226.8
ENSMUST00000124259.8
|
Ggt1
|
gamma-glutamyltransferase 1
|
|
chr5_+_33815466
Show fit
|
13.05 |
ENSMUST00000074849.13
ENSMUST00000079534.11
ENSMUST00000201633.2
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3
|
|
chr17_+_28426752
Show fit
|
12.90 |
ENSMUST00000002327.6
ENSMUST00000233560.2
ENSMUST00000233958.2
ENSMUST00000233170.2
|
Def6
|
differentially expressed in FDCP 6
|
|
chr4_-_154110324
Show fit
|
12.34 |
ENSMUST00000130175.8
ENSMUST00000182151.8
|
Smim1
|
small integral membrane protein 1
|
|
chr11_-_99328969
Show fit
|
12.15 |
ENSMUST00000017743.3
|
Krt20
|
keratin 20
|
|
chr4_-_133695204
Show fit
|
12.06 |
ENSMUST00000100472.10
ENSMUST00000136327.2
|
Hmgn2
|
high mobility group nucleosomal binding domain 2
|
|
chr4_-_154110073
Show fit
|
11.88 |
ENSMUST00000182191.8
ENSMUST00000146543.9
ENSMUST00000146426.2
|
Smim1
|
small integral membrane protein 1
|
|
chr11_+_23206001
Show fit
|
11.81 |
ENSMUST00000020538.13
ENSMUST00000109551.8
ENSMUST00000102870.8
ENSMUST00000102869.8
|
Xpo1
|
exportin 1
|
|
chrX_+_72683020
Show fit
|
11.76 |
ENSMUST00000019701.9
|
Dusp9
|
dual specificity phosphatase 9
|
|
chr4_-_154110383
Show fit
|
11.71 |
ENSMUST00000132541.8
ENSMUST00000143047.8
|
Smim1
|
small integral membrane protein 1
|
|
chr12_+_24758968
Show fit
|
11.08 |
ENSMUST00000154588.2
|
Rrm2
|
ribonucleotide reductase M2
|
|
chr19_-_40576897
Show fit
|
10.87 |
ENSMUST00000025979.13
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1
|
|
chr19_-_40576782
Show fit
|
10.64 |
ENSMUST00000176939.8
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1
|
|
chr8_+_3715747
Show fit
|
10.36 |
ENSMUST00000014118.4
|
Mcemp1
|
mast cell expressed membrane protein 1
|
|
chr4_-_133695264
Show fit
|
9.28 |
ENSMUST00000102553.11
|
Hmgn2
|
high mobility group nucleosomal binding domain 2
|
|
chr5_+_33815910
Show fit
|
9.09 |
ENSMUST00000114426.10
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3
|
|
chr7_+_95860863
Show fit
|
8.87 |
ENSMUST00000107165.8
|
Tenm4
|
teneurin transmembrane protein 4
|
|
chr11_+_96820091
Show fit
|
8.55 |
ENSMUST00000054311.6
ENSMUST00000107636.4
|
Prr15l
|
proline rich 15-like
|
|
chr7_-_44983080
Show fit
|
8.46 |
ENSMUST00000211743.2
ENSMUST00000042194.10
|
Trpm4
|
transient receptor potential cation channel, subfamily M, member 4
|
|
chr11_+_69805005
Show fit
|
8.45 |
ENSMUST00000057884.6
|
Gps2
|
G protein pathway suppressor 2
|
|
chr2_+_84629172
Show fit
|
8.14 |
ENSMUST00000102642.9
ENSMUST00000150325.2
|
Ube2l6
|
ubiquitin-conjugating enzyme E2L 6
|
|
chrX_-_36349055
Show fit
|
7.84 |
ENSMUST00000115231.4
|
Rpl39
|
ribosomal protein L39
|
|
chr19_-_40576817
Show fit
|
7.80 |
ENSMUST00000175932.2
ENSMUST00000176955.8
ENSMUST00000149476.3
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1
|
|
chrX_-_74174524
Show fit
|
7.23 |
ENSMUST00000114091.8
|
Mpp1
|
membrane protein, palmitoylated
|
|
chr14_+_75373766
Show fit
|
6.76 |
ENSMUST00000145303.8
|
Lcp1
|
lymphocyte cytosolic protein 1
|
|
chr5_+_107112186
Show fit
|
6.73 |
ENSMUST00000117196.9
ENSMUST00000031221.12
ENSMUST00000076467.13
|
Cdc7
|
cell division cycle 7 (S. cerevisiae)
|
|
chr5_+_114924106
Show fit
|
6.72 |
ENSMUST00000137519.2
|
Ankrd13a
|
ankyrin repeat domain 13a
|
|
chr17_+_28426831
Show fit
|
6.67 |
ENSMUST00000233264.2
|
Def6
|
differentially expressed in FDCP 6
|
|
chr14_+_75373915
Show fit
|
6.44 |
ENSMUST00000122840.8
|
Lcp1
|
lymphocyte cytosolic protein 1
|
|
chr15_+_89206923
Show fit
|
6.41 |
ENSMUST00000066991.7
|
Adm2
|
adrenomedullin 2
|
|
chr3_-_102871440
Show fit
|
6.37 |
ENSMUST00000058899.13
|
Nr1h5
|
nuclear receptor subfamily 1, group H, member 5
|
|
chr1_+_130659700
Show fit
|
6.35 |
ENSMUST00000039323.8
|
AA986860
|
expressed sequence AA986860
|
|
chr4_+_114945905
Show fit
|
6.29 |
ENSMUST00000171877.8
ENSMUST00000177647.8
ENSMUST00000106548.9
ENSMUST00000030488.3
|
Pdzk1ip1
|
PDZK1 interacting protein 1
|
|
chr17_-_24746911
Show fit
|
6.14 |
ENSMUST00000176652.8
|
Traf7
|
TNF receptor-associated factor 7
|
|
chr10_+_79855454
Show fit
|
6.12 |
ENSMUST00000043311.7
|
Arhgap45
|
Rho GTPase activating protein 45
|
|
chr5_-_30278552
Show fit
|
6.11 |
ENSMUST00000198095.2
ENSMUST00000196872.2
ENSMUST00000026846.11
|
Tyms
|
thymidylate synthase
|
|
chr19_-_34143437
Show fit
|
6.09 |
ENSMUST00000025686.9
|
Ankrd22
|
ankyrin repeat domain 22
|
|
chr17_-_24746804
Show fit
|
6.07 |
ENSMUST00000176353.8
ENSMUST00000176237.8
|
Traf7
|
TNF receptor-associated factor 7
|
|
chr14_-_70945434
Show fit
|
5.98 |
ENSMUST00000228346.2
|
Xpo7
|
exportin 7
|
|
chr8_-_111573401
Show fit
|
5.89 |
ENSMUST00000042012.7
|
Sf3b3
|
splicing factor 3b, subunit 3
|
|
chr17_+_35278011
Show fit
|
5.78 |
ENSMUST00000007255.13
ENSMUST00000174493.8
|
Ddah2
|
dimethylarginine dimethylaminohydrolase 2
|
|
chr11_+_50267808
Show fit
|
5.70 |
ENSMUST00000109142.8
|
Hnrnph1
|
heterogeneous nuclear ribonucleoprotein H1
|
|
chr9_+_50768224
Show fit
|
5.68 |
ENSMUST00000174628.8
ENSMUST00000034560.14
ENSMUST00000114437.9
ENSMUST00000175645.8
ENSMUST00000176349.8
ENSMUST00000176798.8
ENSMUST00000175640.8
|
Ppp2r1b
|
protein phosphatase 2, regulatory subunit A, beta
|
|
chr11_+_48691175
Show fit
|
5.60 |
ENSMUST00000020640.8
|
Rack1
|
receptor for activated C kinase 1
|
|
chrX_+_55824797
Show fit
|
5.51 |
ENSMUST00000114773.10
|
Fhl1
|
four and a half LIM domains 1
|
|
chr5_-_140634773
Show fit
|
5.46 |
ENSMUST00000197452.5
ENSMUST00000042661.8
|
Ttyh3
|
tweety family member 3
|
|
chr2_-_51824937
Show fit
|
5.26 |
ENSMUST00000102767.8
ENSMUST00000102768.8
|
Rbm43
|
RNA binding motif protein 43
|
|
chr8_+_124059414
Show fit
|
5.19 |
ENSMUST00000010298.7
|
Spire2
|
spire type actin nucleation factor 2
|
|
chr2_-_148285450
Show fit
|
5.07 |
ENSMUST00000099269.4
|
Cd93
|
CD93 antigen
|
|
chrX_+_7708034
Show fit
|
4.95 |
ENSMUST00000033494.16
ENSMUST00000115666.8
|
Otud5
|
OTU domain containing 5
|
|
chr7_+_141027763
Show fit
|
4.88 |
ENSMUST00000106003.2
|
Rplp2
|
ribosomal protein, large P2
|
|
chrX_-_162859126
Show fit
|
4.75 |
ENSMUST00000071667.9
|
Siah1b
|
siah E3 ubiquitin protein ligase 1B
|
|
chrX_-_162859095
Show fit
|
4.75 |
ENSMUST00000037928.9
|
Siah1b
|
siah E3 ubiquitin protein ligase 1B
|
|
chr7_+_97480125
Show fit
|
4.69 |
ENSMUST00000206351.2
|
Pak1
|
p21 (RAC1) activated kinase 1
|
|
chr15_-_79976016
Show fit
|
4.61 |
ENSMUST00000185306.3
|
Rpl3
|
ribosomal protein L3
|
|
chr2_+_84966569
Show fit
|
4.56 |
ENSMUST00000057019.9
|
Aplnr
|
apelin receptor
|
|
chr18_+_11766333
Show fit
|
4.54 |
ENSMUST00000115861.9
|
Rbbp8
|
retinoblastoma binding protein 8, endonuclease
|
|
chr7_-_35096133
Show fit
|
4.47 |
ENSMUST00000154597.2
ENSMUST00000032704.12
|
Faap24
|
Fanconi anemia core complex associated protein 24
|
|
chr19_-_8690330
Show fit
|
4.41 |
ENSMUST00000206598.2
|
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2
|
|
chr17_+_34812361
Show fit
|
4.31 |
ENSMUST00000174532.2
|
Pbx2
|
pre B cell leukemia homeobox 2
|
|
chr10_+_13376745
Show fit
|
4.30 |
ENSMUST00000060212.13
ENSMUST00000121465.3
|
Fuca2
|
fucosidase, alpha-L- 2, plasma
|
|
chr6_-_34294377
Show fit
|
4.28 |
ENSMUST00000154655.2
ENSMUST00000102980.11
|
Akr1b3
|
aldo-keto reductase family 1, member B3 (aldose reductase)
|
|
chr19_-_46917661
Show fit
|
4.28 |
ENSMUST00000236727.2
|
Nt5c2
|
5'-nucleotidase, cytosolic II
|
|
chr13_-_76166789
Show fit
|
4.27 |
ENSMUST00000179078.9
ENSMUST00000167271.9
|
Rfesd
|
Rieske (Fe-S) domain containing
|
|
chr12_-_114252202
Show fit
|
4.22 |
ENSMUST00000195124.6
ENSMUST00000103481.3
|
Ighv3-6
|
immunoglobulin heavy variable 3-6
|
|
chr3_-_92493507
Show fit
|
4.13 |
ENSMUST00000194965.6
|
Smcp
|
sperm mitochondria-associated cysteine-rich protein
|
|
chr6_+_5390386
Show fit
|
3.92 |
ENSMUST00000183358.2
|
Asb4
|
ankyrin repeat and SOCS box-containing 4
|
|
chr5_-_34794185
Show fit
|
3.85 |
ENSMUST00000149657.5
|
Mfsd10
|
major facilitator superfamily domain containing 10
|
|
chr19_-_38032006
Show fit
|
3.85 |
ENSMUST00000172095.3
ENSMUST00000041475.16
|
Myof
|
myoferlin
|
|
chr8_+_121215155
Show fit
|
3.77 |
ENSMUST00000034279.16
|
Gse1
|
genetic suppressor element 1, coiled-coil protein
|
|
chr2_-_167852538
Show fit
|
3.72 |
ENSMUST00000099073.3
|
Ripor3
|
RIPOR family member 3
|
|
chr6_-_116693849
Show fit
|
3.64 |
ENSMUST00000056623.13
|
Tmem72
|
transmembrane protein 72
|
|
chr1_-_149836974
Show fit
|
3.62 |
ENSMUST00000190507.2
ENSMUST00000070200.15
|
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent)
|
|
chr12_-_91712783
Show fit
|
3.53 |
ENSMUST00000166967.2
|
Ston2
|
stonin 2
|
|
chr15_-_103446354
Show fit
|
3.51 |
ENSMUST00000023133.8
|
Ppp1r1a
|
protein phosphatase 1, regulatory inhibitor subunit 1A
|
|
chr7_+_101619019
Show fit
|
3.45 |
ENSMUST00000084852.13
|
Numa1
|
nuclear mitotic apparatus protein 1
|
|
chr16_-_19079594
Show fit
|
3.43 |
ENSMUST00000103752.3
ENSMUST00000197518.2
|
Iglv2
|
immunoglobulin lambda variable 2
|
|
chr19_-_38031774
Show fit
|
3.33 |
ENSMUST00000226068.2
|
Myof
|
myoferlin
|
|
chr5_-_129864202
Show fit
|
3.27 |
ENSMUST00000136507.4
|
Psph
|
phosphoserine phosphatase
|
|
chr14_+_30201569
Show fit
|
3.13 |
ENSMUST00000022535.9
ENSMUST00000223658.2
|
Dcp1a
|
decapping mRNA 1A
|
|
chr12_+_78908466
Show fit
|
3.06 |
ENSMUST00000071230.8
|
Eif2s1
|
eukaryotic translation initiation factor 2, subunit 1 alpha
|
|
chr5_-_142892457
Show fit
|
3.04 |
ENSMUST00000167721.8
ENSMUST00000163829.2
ENSMUST00000100497.11
|
Actb
|
actin, beta
|
|
chr4_-_139079842
Show fit
|
3.04 |
ENSMUST00000102503.10
|
Mrto4
|
mRNA turnover 4, ribosome maturation factor
|
|
chr9_+_123902143
Show fit
|
3.02 |
ENSMUST00000168841.3
ENSMUST00000055918.7
|
Ccr2
|
chemokine (C-C motif) receptor 2
|
|
chr11_-_102207486
Show fit
|
3.02 |
ENSMUST00000146896.9
ENSMUST00000079589.11
|
Ubtf
|
upstream binding transcription factor, RNA polymerase I
|
|
chr11_+_75422516
Show fit
|
3.01 |
ENSMUST00000149727.8
ENSMUST00000108433.8
ENSMUST00000042561.14
ENSMUST00000143035.8
|
Slc43a2
|
solute carrier family 43, member 2
|
|
chr3_+_58433236
Show fit
|
2.96 |
ENSMUST00000029387.15
|
Eif2a
|
eukaryotic translation initiation factor 2A
|
|
chr8_+_70275079
Show fit
|
2.94 |
ENSMUST00000164890.8
ENSMUST00000034325.6
ENSMUST00000238452.2
|
Lpar2
|
lysophosphatidic acid receptor 2
|
|
chrX_+_7708295
Show fit
|
2.91 |
ENSMUST00000115667.10
ENSMUST00000115668.10
ENSMUST00000115665.2
|
Otud5
|
OTU domain containing 5
|
|
chr8_-_61407760
Show fit
|
2.87 |
ENSMUST00000110302.8
|
Clcn3
|
chloride channel, voltage-sensitive 3
|
|
chr5_-_134717443
Show fit
|
2.83 |
ENSMUST00000015137.10
|
Limk1
|
LIM-domain containing, protein kinase
|
|
chr11_-_102207516
Show fit
|
2.81 |
ENSMUST00000107115.8
ENSMUST00000128016.2
|
Ubtf
|
upstream binding transcription factor, RNA polymerase I
|
|
chr1_-_60137263
Show fit
|
2.80 |
ENSMUST00000143342.8
|
Wdr12
|
WD repeat domain 12
|
|
chr12_-_113700190
Show fit
|
2.77 |
ENSMUST00000103452.3
ENSMUST00000192264.2
|
Ighv5-9-1
|
immunoglobulin heavy variable 5-9-1
|
|
chr7_+_35096470
Show fit
|
2.73 |
ENSMUST00000079414.12
|
Cep89
|
centrosomal protein 89
|
|
chr11_-_70860778
Show fit
|
2.70 |
ENSMUST00000108530.2
ENSMUST00000035283.11
ENSMUST00000108531.8
|
Nup88
|
nucleoporin 88
|
|
chr11_-_45846291
Show fit
|
2.67 |
ENSMUST00000011398.13
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae)
|
|
chr8_+_34089597
Show fit
|
2.67 |
ENSMUST00000009774.11
|
Ppp2cb
|
protein phosphatase 2 (formerly 2A), catalytic subunit, beta isoform
|
|
chr1_-_52230062
Show fit
|
2.64 |
ENSMUST00000156887.8
ENSMUST00000129107.2
|
Gls
|
glutaminase
|
|
chr11_-_82719850
Show fit
|
2.62 |
ENSMUST00000021036.13
ENSMUST00000074515.11
ENSMUST00000103218.3
|
Rffl
|
ring finger and FYVE like domain containing protein
|
|
chr19_-_53932581
Show fit
|
2.55 |
ENSMUST00000236885.2
ENSMUST00000236098.2
ENSMUST00000236370.2
|
Bbip1
|
BBSome interacting protein 1
|
|
chr6_-_39534765
Show fit
|
2.47 |
ENSMUST00000036877.10
ENSMUST00000154149.2
|
Dennd2a
|
DENN/MADD domain containing 2A
|
|
chr12_-_114286421
Show fit
|
2.43 |
ENSMUST00000103483.3
|
Ighv3-8
|
immunoglobulin heavy variable V3-8
|
|
chr4_-_139079609
Show fit
|
2.41 |
ENSMUST00000030513.13
ENSMUST00000155257.8
|
Mrto4
|
mRNA turnover 4, ribosome maturation factor
|
|
chr3_+_84859453
Show fit
|
2.41 |
ENSMUST00000029727.8
|
Fbxw7
|
F-box and WD-40 domain protein 7
|
|
chr11_-_8614497
Show fit
|
2.41 |
ENSMUST00000020695.13
|
Tns3
|
tensin 3
|
|
chr1_+_55091922
Show fit
|
2.38 |
ENSMUST00000087617.11
ENSMUST00000027125.7
|
Coq10b
|
coenzyme Q10B
|
|
chr4_-_129334593
Show fit
|
2.35 |
ENSMUST00000053042.6
ENSMUST00000106046.8
|
Zbtb8b
|
zinc finger and BTB domain containing 8b
|
|
chr16_-_3725515
Show fit
|
2.33 |
ENSMUST00000177221.2
ENSMUST00000177323.8
|
1700037C18Rik
|
RIKEN cDNA 1700037C18 gene
|
|
chr3_+_146156220
Show fit
|
2.27 |
ENSMUST00000061937.13
ENSMUST00000029840.4
|
Ctbs
|
chitobiase
|
|
chr3_+_122522592
Show fit
|
2.24 |
ENSMUST00000066728.10
|
Pde5a
|
phosphodiesterase 5A, cGMP-specific
|
|
chr19_-_45738002
Show fit
|
2.17 |
ENSMUST00000070215.8
|
Npm3
|
nucleoplasmin 3
|
|
chr6_-_142517340
Show fit
|
2.14 |
ENSMUST00000203945.3
|
Kcnj8
|
potassium inwardly-rectifying channel, subfamily J, member 8
|
|
chr5_+_129864044
Show fit
|
2.06 |
ENSMUST00000201414.5
|
Cct6a
|
chaperonin containing Tcp1, subunit 6a (zeta)
|
|
chr9_-_67739607
Show fit
|
1.98 |
ENSMUST00000054500.7
|
C2cd4a
|
C2 calcium-dependent domain containing 4A
|
|
chr19_-_24178000
Show fit
|
1.92 |
ENSMUST00000233658.3
|
Tjp2
|
tight junction protein 2
|
|
chr19_+_29344846
Show fit
|
1.92 |
ENSMUST00000016640.8
|
Cd274
|
CD274 antigen
|
|
chr18_+_61058684
Show fit
|
1.91 |
ENSMUST00000102888.10
ENSMUST00000025519.11
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr19_-_29344694
Show fit
|
1.84 |
ENSMUST00000138051.2
|
Plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein
|
|
chr19_-_10847121
Show fit
|
1.84 |
ENSMUST00000120524.2
ENSMUST00000025645.14
|
Tmem132a
|
transmembrane protein 132A
|
|
chr3_+_123240562
Show fit
|
1.78 |
ENSMUST00000029603.10
|
Prss12
|
protease, serine 12 neurotrypsin (motopsin)
|
|
chr11_-_61065846
Show fit
|
1.76 |
ENSMUST00000041683.9
|
Usp22
|
ubiquitin specific peptidase 22
|
|
chr5_-_100648487
Show fit
|
1.69 |
ENSMUST00000239512.1
|
LIN54
|
lin-54 homolog (C. elegans)
|
|
chr11_-_8614667
Show fit
|
1.61 |
ENSMUST00000239111.2
|
Tns3
|
tensin 3
|
|
chr4_-_133329479
Show fit
|
1.57 |
ENSMUST00000057311.4
|
Sfn
|
stratifin
|
|
chr2_+_32460868
Show fit
|
1.57 |
ENSMUST00000140592.8
ENSMUST00000028151.7
|
Dpm2
|
dolichol-phosphate (beta-D) mannosyltransferase 2
|
|
chr13_+_51562675
Show fit
|
1.56 |
ENSMUST00000087978.5
|
S1pr3
|
sphingosine-1-phosphate receptor 3
|
|
chr15_-_11905697
Show fit
|
1.53 |
ENSMUST00000066529.5
ENSMUST00000228603.2
|
Npr3
|
natriuretic peptide receptor 3
|
|
chr16_+_43960183
Show fit
|
1.50 |
ENSMUST00000159514.8
ENSMUST00000161326.8
ENSMUST00000063520.15
ENSMUST00000063542.8
|
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr11_-_95805710
Show fit
|
1.46 |
ENSMUST00000038343.7
|
B4galnt2
|
beta-1,4-N-acetyl-galactosaminyl transferase 2
|
|
chr14_-_54651442
Show fit
|
1.45 |
ENSMUST00000227334.2
|
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7
|
|
chr2_-_102016717
Show fit
|
1.42 |
ENSMUST00000058790.12
|
Ldlrad3
|
low density lipoprotein receptor class A domain containing 3
|
|
chrX_-_20816841
Show fit
|
1.40 |
ENSMUST00000009550.14
|
Elk1
|
ELK1, member of ETS oncogene family
|
|
chr6_+_67838100
Show fit
|
1.36 |
ENSMUST00000200586.2
ENSMUST00000103309.3
|
Igkv17-127
|
immunoglobulin kappa variable 17-127
|
|
chrX_+_151016224
Show fit
|
1.36 |
ENSMUST00000112588.9
ENSMUST00000082177.13
|
Kdm5c
|
lysine (K)-specific demethylase 5C
|
|
chr19_-_60849828
Show fit
|
1.35 |
ENSMUST00000135808.8
|
Sfxn4
|
sideroflexin 4
|
|
chr2_-_102016665
Show fit
|
1.34 |
ENSMUST00000111222.2
|
Ldlrad3
|
low density lipoprotein receptor class A domain containing 3
|
|
chr7_-_7139723
Show fit
|
1.33 |
ENSMUST00000211240.2
ENSMUST00000032622.9
|
Zfp773
|
zinc finger protein 773
|
|
chr19_+_6887059
Show fit
|
1.31 |
ENSMUST00000088257.14
ENSMUST00000116551.10
|
Trmt112
|
tRNA methyltransferase 11-2
|
|
chr9_+_35334878
Show fit
|
1.30 |
ENSMUST00000154652.8
|
Cdon
|
cell adhesion molecule-related/down-regulated by oncogenes
|
|
chr4_-_132149704
Show fit
|
1.27 |
ENSMUST00000152271.8
ENSMUST00000084170.12
|
Phactr4
|
phosphatase and actin regulator 4
|
|
chr4_+_118478357
Show fit
|
1.26 |
ENSMUST00000147373.2
|
Ebna1bp2
|
EBNA1 binding protein 2
|
|
chr3_-_95014090
Show fit
|
1.23 |
ENSMUST00000005768.8
ENSMUST00000107232.9
ENSMUST00000107236.9
|
Pip5k1a
|
phosphatidylinositol-4-phosphate 5-kinase, type 1 alpha
|
|
chr4_+_109533753
Show fit
|
1.22 |
ENSMUST00000102724.5
|
Faf1
|
Fas-associated factor 1
|
|
chr15_-_78058214
Show fit
|
1.22 |
ENSMUST00000016781.8
|
Ift27
|
intraflagellar transport 27
|
|
chrX_+_52001108
Show fit
|
1.20 |
ENSMUST00000078944.13
ENSMUST00000101587.10
ENSMUST00000154864.4
|
Phf6
|
PHD finger protein 6
|
|
chr3_-_143908111
Show fit
|
1.16 |
ENSMUST00000121796.8
|
Lmo4
|
LIM domain only 4
|
|
chr14_-_22039543
Show fit
|
1.15 |
ENSMUST00000043409.9
|
Zfp503
|
zinc finger protein 503
|
|
chr4_-_49593875
Show fit
|
1.08 |
ENSMUST00000151542.2
|
Pgap4
|
post-GPI attachment to proteins GalNAc transferase 4
|
|
chr9_-_65734826
Show fit
|
1.07 |
ENSMUST00000159109.2
|
Zfp609
|
zinc finger protein 609
|
|
chr11_+_3845221
Show fit
|
1.06 |
ENSMUST00000109996.8
ENSMUST00000055931.5
|
Dusp18
|
dual specificity phosphatase 18
|
|
chr10_+_69048506
Show fit
|
1.05 |
ENSMUST00000167384.8
|
Rhobtb1
|
Rho-related BTB domain containing 1
|
|
chr10_+_79766254
Show fit
|
1.02 |
ENSMUST00000131118.2
|
Arid3a
|
AT rich interactive domain 3A (BRIGHT-like)
|
|
chr3_+_95018324
Show fit
|
1.01 |
ENSMUST00000009102.9
|
Vps72
|
vacuolar protein sorting 72
|
|
chr13_+_95112852
Show fit
|
1.00 |
ENSMUST00000160115.8
ENSMUST00000222995.2
ENSMUST00000160801.8
ENSMUST00000056512.14
|
Wdr41
|
WD repeat domain 41
|
|
chr11_-_45845992
Show fit
|
0.99 |
ENSMUST00000109254.2
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae)
|
|
chr17_+_32671689
Show fit
|
0.97 |
ENSMUST00000237491.2
|
Cyp4f39
|
cytochrome P450, family 4, subfamily f, polypeptide 39
|
|
chr3_-_143908060
Show fit
|
0.93 |
ENSMUST00000121112.6
|
Lmo4
|
LIM domain only 4
|
|
chr9_+_108368032
Show fit
|
0.90 |
ENSMUST00000166103.9
ENSMUST00000085044.14
ENSMUST00000193678.6
ENSMUST00000178075.8
|
Usp19
|
ubiquitin specific peptidase 19
|
|
chr19_-_55229668
Show fit
|
0.89 |
ENSMUST00000069183.8
|
Gucy2g
|
guanylate cyclase 2g
|
|
chr12_-_114330574
Show fit
|
0.87 |
ENSMUST00000103485.3
|
Ighv12-3
|
immunoglobulin heavy variable V12-3
|
|
chr2_+_156562956
Show fit
|
0.87 |
ENSMUST00000109566.9
ENSMUST00000146412.9
ENSMUST00000177013.8
ENSMUST00000171030.9
|
Dlgap4
|
DLG associated protein 4
|
|
chr14_-_21898992
Show fit
|
0.84 |
ENSMUST00000124549.9
|
Comtd1
|
catechol-O-methyltransferase domain containing 1
|
|
chr13_+_119599287
Show fit
|
0.83 |
ENSMUST00000026519.10
ENSMUST00000225186.2
ENSMUST00000224081.2
ENSMUST00000224312.2
|
4833420G17Rik
|
RIKEN cDNA 4833420G17 gene
|
|
chr18_+_65715156
Show fit
|
0.82 |
ENSMUST00000237553.2
ENSMUST00000237712.2
ENSMUST00000182684.8
|
Zfp532
|
zinc finger protein 532
|
|
chrX_+_163763588
Show fit
|
0.80 |
ENSMUST00000167446.8
ENSMUST00000057150.8
|
Fancb
|
Fanconi anemia, complementation group B
|
|
chr14_+_79718604
Show fit
|
0.80 |
ENSMUST00000040131.13
|
Elf1
|
E74-like factor 1
|
|
chr13_-_76166643
Show fit
|
0.80 |
ENSMUST00000050997.2
|
Rfesd
|
Rieske (Fe-S) domain containing
|
|
chr4_-_149858694
Show fit
|
0.80 |
ENSMUST00000105686.3
|
Slc25a33
|
solute carrier family 25, member 33
|
|
chr18_+_61688378
Show fit
|
0.79 |
ENSMUST00000165721.8
ENSMUST00000115246.9
ENSMUST00000166990.8
ENSMUST00000163205.8
ENSMUST00000170862.8
|
Csnk1a1
|
casein kinase 1, alpha 1
|
|
chr3_-_95014218
Show fit
|
0.76 |
ENSMUST00000107233.9
|
Pip5k1a
|
phosphatidylinositol-4-phosphate 5-kinase, type 1 alpha
|
|
chr18_+_37880027
Show fit
|
0.75 |
ENSMUST00000193404.2
|
Pcdhga10
|
protocadherin gamma subfamily A, 10
|
|
chr11_+_103857541
Show fit
|
0.71 |
ENSMUST00000057921.10
ENSMUST00000063347.12
|
Arf2
|
ADP-ribosylation factor 2
|
|
chr3_+_138233004
Show fit
|
0.71 |
ENSMUST00000196990.5
ENSMUST00000200239.5
ENSMUST00000200100.2
|
Eif4e
|
eukaryotic translation initiation factor 4E
|
|
chr19_-_8906686
Show fit
|
0.70 |
ENSMUST00000096242.5
|
Rom1
|
rod outer segment membrane protein 1
|
|
chr18_+_37869950
Show fit
|
0.68 |
ENSMUST00000091935.7
|
Pcdhga9
|
protocadherin gamma subfamily A, 9
|
|
chr10_-_129962924
Show fit
|
0.66 |
ENSMUST00000074161.2
|
Olfr824
|
olfactory receptor 824
|
|
chr12_-_70160491
Show fit
|
0.62 |
ENSMUST00000222237.2
|
Nin
|
ninein
|
|
chr4_-_116024788
Show fit
|
0.58 |
ENSMUST00000030465.10
ENSMUST00000143426.2
|
Tspan1
|
tetraspanin 1
|
|
chr11_+_4852212
Show fit
|
0.54 |
ENSMUST00000142543.3
|
Thoc5
|
THO complex 5
|
|
chr2_+_16361582
Show fit
|
0.52 |
ENSMUST00000114703.10
|
Plxdc2
|
plexin domain containing 2
|
|
chr9_+_15150341
Show fit
|
0.45 |
ENSMUST00000034413.8
|
Vstm5
|
V-set and transmembrane domain containing 5
|
|
chr17_+_37148015
Show fit
|
0.45 |
ENSMUST00000179968.8
ENSMUST00000130367.8
ENSMUST00000053434.15
ENSMUST00000130801.8
ENSMUST00000144182.8
ENSMUST00000123715.8
|
Trim26
|
tripartite motif-containing 26
|
|
chr2_-_132420047
Show fit
|
0.43 |
ENSMUST00000028822.14
|
Gpcpd1
|
glycerophosphocholine phosphodiesterase 1
|
|
chr14_-_45096827
Show fit
|
0.43 |
ENSMUST00000095959.2
|
Ptgdr
|
prostaglandin D receptor
|
|
chr16_-_32630847
Show fit
|
0.43 |
ENSMUST00000179384.3
|
Smbd1
|
somatomedin B domain containing 1
|
|
chr3_-_41696906
Show fit
|
0.42 |
ENSMUST00000026866.15
|
Sclt1
|
sodium channel and clathrin linker 1
|
|
chr10_-_127587576
Show fit
|
0.42 |
ENSMUST00000079692.6
|
Gpr182
|
G protein-coupled receptor 182
|
|
chr16_+_20492267
Show fit
|
0.42 |
ENSMUST00000115460.8
|
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1
|
|
chr3_+_90201388
Show fit
|
0.42 |
ENSMUST00000199607.5
|
Gatad2b
|
GATA zinc finger domain containing 2B
|