Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Tlx1
Z-value: 2.49
Transcription factors associated with Tlx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tlx1
|
ENSMUSG00000025215.11 | Tlx1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tlx1 | mm39_v1_chr19_+_45139908_45140034 | -0.14 | 4.3e-01 | Click! |
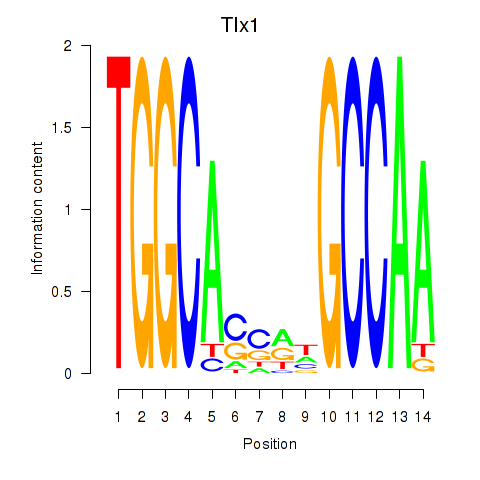
Activity profile of Tlx1 motif
Sorted Z-values of Tlx1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tlx1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_55152002 | 28.07 |
ENSMUST00000003569.6
|
Inmt
|
indolethylamine N-methyltransferase |
| chr9_-_57590926 | 21.48 |
ENSMUST00000034860.5
|
Cyp1a2
|
cytochrome P450, family 1, subfamily a, polypeptide 2 |
| chr7_+_119125426 | 19.43 |
ENSMUST00000066465.3
|
Acsm5
|
acyl-CoA synthetase medium-chain family member 5 |
| chr7_+_119125546 | 19.35 |
ENSMUST00000207387.2
ENSMUST00000207813.2 |
Acsm5
|
acyl-CoA synthetase medium-chain family member 5 |
| chr7_+_119125443 | 18.83 |
ENSMUST00000207440.2
|
Acsm5
|
acyl-CoA synthetase medium-chain family member 5 |
| chr7_+_26534730 | 16.95 |
ENSMUST00000005685.15
|
Cyp2a5
|
cytochrome P450, family 2, subfamily a, polypeptide 5 |
| chr7_-_97066937 | 15.03 |
ENSMUST00000043077.8
|
Thrsp
|
thyroid hormone responsive |
| chr17_-_56428968 | 10.56 |
ENSMUST00000041357.9
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr10_-_39901249 | 9.76 |
ENSMUST00000163705.3
|
Mfsd4b1
|
major facilitator superfamily domain containing 4B1 |
| chr14_+_66208253 | 9.23 |
ENSMUST00000138191.8
|
Clu
|
clusterin |
| chr10_+_128089965 | 9.19 |
ENSMUST00000060782.5
ENSMUST00000218722.2 |
Apon
|
apolipoprotein N |
| chr10_-_81127057 | 8.49 |
ENSMUST00000045744.7
|
Tjp3
|
tight junction protein 3 |
| chr14_+_66208059 | 8.35 |
ENSMUST00000127387.8
|
Clu
|
clusterin |
| chr14_+_51328534 | 7.97 |
ENSMUST00000022428.13
ENSMUST00000171688.9 |
Rnase4
Ang
|
ribonuclease, RNase A family 4 angiogenin, ribonuclease, RNase A family, 5 |
| chr14_+_66208498 | 7.89 |
ENSMUST00000128539.8
|
Clu
|
clusterin |
| chr11_+_115353290 | 7.73 |
ENSMUST00000106532.4
ENSMUST00000092445.12 ENSMUST00000153466.2 |
Slc16a5
|
solute carrier family 16 (monocarboxylic acid transporters), member 5 |
| chr19_+_32597379 | 7.71 |
ENSMUST00000236290.2
ENSMUST00000025833.7 |
Papss2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr14_+_66208613 | 7.58 |
ENSMUST00000144619.2
|
Clu
|
clusterin |
| chr7_+_26006594 | 7.39 |
ENSMUST00000098657.5
|
Cyp2a4
|
cytochrome P450, family 2, subfamily a, polypeptide 4 |
| chr5_+_146016064 | 7.12 |
ENSMUST00000035571.10
|
Cyp3a59
|
cytochrome P450, family 3, subfamily a, polypeptide 59 |
| chr13_-_42001075 | 6.87 |
ENSMUST00000179758.8
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr7_-_12732067 | 6.70 |
ENSMUST00000032539.14
ENSMUST00000120903.8 |
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr13_-_42000958 | 6.42 |
ENSMUST00000072012.10
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr13_-_42001102 | 6.21 |
ENSMUST00000121404.8
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr9_+_77824646 | 6.02 |
ENSMUST00000034904.14
|
Elovl5
|
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
| chr16_-_18245352 | 5.75 |
ENSMUST00000000335.12
|
Comt
|
catechol-O-methyltransferase |
| chr2_+_118998235 | 5.62 |
ENSMUST00000057454.4
|
Gchfr
|
GTP cyclohydrolase I feedback regulator |
| chr7_-_12731594 | 5.51 |
ENSMUST00000133977.3
|
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr17_+_32904629 | 5.34 |
ENSMUST00000008801.7
|
Cyp4f15
|
cytochrome P450, family 4, subfamily f, polypeptide 15 |
| chr17_+_32904601 | 5.28 |
ENSMUST00000168171.8
|
Cyp4f15
|
cytochrome P450, family 4, subfamily f, polypeptide 15 |
| chr5_-_77262968 | 5.27 |
ENSMUST00000081964.7
|
Hopx
|
HOP homeobox |
| chr19_-_46661501 | 5.21 |
ENSMUST00000236174.2
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr19_-_46661321 | 5.18 |
ENSMUST00000026012.8
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr4_-_57916283 | 4.76 |
ENSMUST00000063816.6
|
D630039A03Rik
|
RIKEN cDNA D630039A03 gene |
| chr3_-_63872189 | 4.61 |
ENSMUST00000029402.15
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr1_-_162687254 | 4.54 |
ENSMUST00000131058.8
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr1_-_162687369 | 4.53 |
ENSMUST00000193078.6
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr11_-_75313412 | 4.50 |
ENSMUST00000138661.8
ENSMUST00000000769.14 |
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
| chr11_-_75313350 | 4.42 |
ENSMUST00000125982.2
ENSMUST00000137103.8 |
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
| chr7_-_126275529 | 4.24 |
ENSMUST00000106372.11
ENSMUST00000155419.3 ENSMUST00000106373.9 |
Sult1a1
|
sulfotransferase family 1A, phenol-preferring, member 1 |
| chr2_-_91025441 | 4.13 |
ENSMUST00000002177.9
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr5_+_115061293 | 4.06 |
ENSMUST00000031540.11
ENSMUST00000112143.4 |
Oasl1
|
2'-5' oligoadenylate synthetase-like 1 |
| chr17_-_32639936 | 4.06 |
ENSMUST00000170392.9
ENSMUST00000237165.2 ENSMUST00000235892.2 ENSMUST00000114455.3 |
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr3_+_123061094 | 4.02 |
ENSMUST00000047923.12
ENSMUST00000200333.2 |
Sec24d
|
Sec24 related gene family, member D (S. cerevisiae) |
| chr10_-_76949762 | 4.01 |
ENSMUST00000072755.12
|
Col18a1
|
collagen, type XVIII, alpha 1 |
| chr13_+_12580743 | 3.98 |
ENSMUST00000221560.2
ENSMUST00000071973.8 |
Ero1b
|
endoplasmic reticulum oxidoreductase 1 beta |
| chr10_-_76949510 | 3.96 |
ENSMUST00000105409.8
|
Col18a1
|
collagen, type XVIII, alpha 1 |
| chr2_+_26969384 | 3.83 |
ENSMUST00000091233.7
|
Adamtsl2
|
ADAMTS-like 2 |
| chr3_-_63872079 | 3.79 |
ENSMUST00000161659.8
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr1_+_133173826 | 3.58 |
ENSMUST00000105082.9
ENSMUST00000038295.15 |
Plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr3_+_57643477 | 3.46 |
ENSMUST00000041826.14
ENSMUST00000198510.5 ENSMUST00000200497.5 ENSMUST00000198214.5 ENSMUST00000200600.5 ENSMUST00000198249.5 ENSMUST00000199041.2 |
Rnf13
|
ring finger protein 13 |
| chr15_-_82291372 | 3.34 |
ENSMUST00000230198.2
ENSMUST00000230248.2 ENSMUST00000072776.5 ENSMUST00000229911.2 |
Cyp2d10
|
cytochrome P450, family 2, subfamily d, polypeptide 10 |
| chr4_-_129121676 | 3.28 |
ENSMUST00000106051.8
|
C77080
|
expressed sequence C77080 |
| chr3_-_88162012 | 3.21 |
ENSMUST00000171887.4
|
Rhbg
|
Rhesus blood group-associated B glycoprotein |
| chr4_-_138095277 | 3.19 |
ENSMUST00000030535.4
|
Cda
|
cytidine deaminase |
| chr2_-_91025208 | 3.16 |
ENSMUST00000111355.8
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr4_+_139350152 | 3.16 |
ENSMUST00000039818.10
|
Aldh4a1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr11_+_96920956 | 3.13 |
ENSMUST00000153482.2
|
Scrn2
|
secernin 2 |
| chr2_-_91025380 | 3.04 |
ENSMUST00000111356.8
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr11_+_96920751 | 3.02 |
ENSMUST00000021249.11
|
Scrn2
|
secernin 2 |
| chr3_-_107838895 | 3.02 |
ENSMUST00000133947.9
ENSMUST00000124215.2 ENSMUST00000106688.8 ENSMUST00000106687.9 |
Gstm7
|
glutathione S-transferase, mu 7 |
| chr9_+_106325860 | 2.90 |
ENSMUST00000185527.3
|
Abhd14b
|
abhydrolase domain containing 14b |
| chr9_+_106324952 | 2.87 |
ENSMUST00000215475.2
ENSMUST00000187106.7 ENSMUST00000190167.7 |
Abhd14b
|
abhydrolase domain containing 14b |
| chr2_-_130506484 | 2.81 |
ENSMUST00000089559.11
|
Ddrgk1
|
DDRGK domain containing 1 |
| chr10_-_127457001 | 2.76 |
ENSMUST00000049149.15
|
Lrp1
|
low density lipoprotein receptor-related protein 1 |
| chr15_+_81686622 | 2.75 |
ENSMUST00000109553.10
|
Tef
|
thyrotroph embryonic factor |
| chr2_-_91025492 | 2.69 |
ENSMUST00000111354.2
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr3_+_81904229 | 2.66 |
ENSMUST00000029641.10
|
Asic5
|
acid-sensing (proton-gated) ion channel family member 5 |
| chr19_-_3962733 | 2.66 |
ENSMUST00000075092.8
ENSMUST00000235847.2 ENSMUST00000235301.2 ENSMUST00000237341.2 |
Ndufs8
|
NADH:ubiquinone oxidoreductase core subunit S8 |
| chr9_-_106315518 | 2.65 |
ENSMUST00000024031.13
ENSMUST00000190972.3 |
Acy1
|
aminoacylase 1 |
| chr17_-_24428351 | 2.61 |
ENSMUST00000024931.6
|
Ntn3
|
netrin 3 |
| chr7_-_80053063 | 2.60 |
ENSMUST00000147150.2
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr6_-_115228800 | 2.57 |
ENSMUST00000205131.2
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr11_-_77784922 | 2.51 |
ENSMUST00000017597.5
|
Pipox
|
pipecolic acid oxidase |
| chr14_+_29730931 | 2.47 |
ENSMUST00000067620.12
|
Chdh
|
choline dehydrogenase |
| chr8_-_96161414 | 2.44 |
ENSMUST00000211908.2
|
Cfap20
|
cilia and flagella associated protein 20 |
| chr13_+_12580772 | 2.42 |
ENSMUST00000220811.2
|
Ero1b
|
endoplasmic reticulum oxidoreductase 1 beta |
| chr10_-_89369432 | 2.40 |
ENSMUST00000105297.2
|
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr3_-_107839133 | 2.39 |
ENSMUST00000004137.11
|
Gstm7
|
glutathione S-transferase, mu 7 |
| chr6_-_33037107 | 2.36 |
ENSMUST00000115091.2
ENSMUST00000127666.8 |
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr10_-_127456791 | 2.36 |
ENSMUST00000118455.2
ENSMUST00000121829.8 |
Lrp1
|
low density lipoprotein receptor-related protein 1 |
| chr1_+_87501721 | 2.33 |
ENSMUST00000166259.8
ENSMUST00000172222.8 ENSMUST00000163606.8 |
Neu2
|
neuraminidase 2 |
| chr6_-_33037191 | 2.32 |
ENSMUST00000066379.11
|
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr11_+_109376432 | 2.30 |
ENSMUST00000106697.8
|
Arsg
|
arylsulfatase G |
| chr3_-_85653573 | 2.25 |
ENSMUST00000118408.8
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr6_+_48570817 | 2.24 |
ENSMUST00000154010.8
ENSMUST00000009420.15 ENSMUST00000163452.7 ENSMUST00000118229.2 ENSMUST00000135151.3 |
Repin1
|
replication initiator 1 |
| chr16_+_4501934 | 2.20 |
ENSMUST00000060067.12
ENSMUST00000115854.4 ENSMUST00000229529.2 |
Dnaja3
|
DnaJ heat shock protein family (Hsp40) member A3 |
| chrX_+_72271878 | 2.19 |
ENSMUST00000105111.4
|
F8a
|
factor 8-associated gene A |
| chr15_-_74869684 | 2.16 |
ENSMUST00000190188.2
ENSMUST00000189068.7 ENSMUST00000186526.7 ENSMUST00000187171.2 ENSMUST00000187994.7 |
Ly6a
|
lymphocyte antigen 6 complex, locus A |
| chr12_+_95658987 | 2.10 |
ENSMUST00000057324.4
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr6_+_55428963 | 2.08 |
ENSMUST00000070736.12
ENSMUST00000070756.12 ENSMUST00000166962.8 |
Adcyap1r1
|
adenylate cyclase activating polypeptide 1 receptor 1 |
| chr4_-_115504907 | 2.00 |
ENSMUST00000102707.10
|
Cyp4b1
|
cytochrome P450, family 4, subfamily b, polypeptide 1 |
| chrX_-_71699740 | 1.93 |
ENSMUST00000055966.13
|
Gabra3
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 3 |
| chr7_-_127490139 | 1.88 |
ENSMUST00000205300.2
ENSMUST00000121394.3 |
Prss53
|
protease, serine 53 |
| chr4_-_103072343 | 1.87 |
ENSMUST00000150285.8
|
Slc35d1
|
solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1 |
| chr6_-_115229128 | 1.86 |
ENSMUST00000032462.9
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr9_-_20887967 | 1.83 |
ENSMUST00000214218.2
|
S1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr11_+_28803188 | 1.82 |
ENSMUST00000020759.12
|
Efemp1
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 1 |
| chr11_+_4833186 | 1.78 |
ENSMUST00000139737.2
|
Nipsnap1
|
nipsnap homolog 1 |
| chr10_+_60925130 | 1.76 |
ENSMUST00000020298.8
|
Pcbd1
|
pterin 4 alpha carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 1 |
| chr5_+_124621521 | 1.75 |
ENSMUST00000111453.2
|
Snrnp35
|
small nuclear ribonucleoprotein 35 (U11/U12) |
| chr14_-_63415235 | 1.72 |
ENSMUST00000054963.10
|
Fdft1
|
farnesyl diphosphate farnesyl transferase 1 |
| chr14_+_14296748 | 1.71 |
ENSMUST00000022268.10
|
Pdhb
|
pyruvate dehydrogenase (lipoamide) beta |
| chr17_-_12726591 | 1.70 |
ENSMUST00000024595.4
|
Slc22a3
|
solute carrier family 22 (organic cation transporter), member 3 |
| chr4_+_59581617 | 1.67 |
ENSMUST00000107528.8
|
Hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr1_+_176642226 | 1.66 |
ENSMUST00000056773.15
ENSMUST00000027785.15 |
Sdccag8
|
serologically defined colon cancer antigen 8 |
| chr10_+_60925108 | 1.66 |
ENSMUST00000218005.2
|
Pcbd1
|
pterin 4 alpha carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 1 |
| chrX_+_93278526 | 1.64 |
ENSMUST00000113908.8
ENSMUST00000113916.10 |
Klhl15
|
kelch-like 15 |
| chr1_+_167177545 | 1.58 |
ENSMUST00000028004.11
|
Aldh9a1
|
aldehyde dehydrogenase 9, subfamily A1 |
| chr4_+_59581557 | 1.56 |
ENSMUST00000030078.12
|
Hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr2_-_26012751 | 1.53 |
ENSMUST00000140993.2
ENSMUST00000028300.6 |
Nacc2
|
nucleus accumbens associated 2, BEN and BTB (POZ) domain containing |
| chr6_+_17306414 | 1.52 |
ENSMUST00000150901.2
|
Cav1
|
caveolin 1, caveolae protein |
| chr17_+_6157154 | 1.50 |
ENSMUST00000149756.8
|
Tulp4
|
tubby like protein 4 |
| chr11_-_120604551 | 1.50 |
ENSMUST00000106154.8
ENSMUST00000106155.4 ENSMUST00000055424.13 ENSMUST00000026137.8 |
Cenpx
|
centromere protein X |
| chrX_+_93278588 | 1.49 |
ENSMUST00000096369.10
ENSMUST00000113911.9 |
Klhl15
|
kelch-like 15 |
| chr13_-_74642055 | 1.48 |
ENSMUST00000202645.4
ENSMUST00000221173.2 |
Zfp825
|
zinc finger protein 825 |
| chr2_+_164674782 | 1.48 |
ENSMUST00000103093.10
|
Ctsa
|
cathepsin A |
| chr1_-_87501548 | 1.47 |
ENSMUST00000068681.12
|
Ngef
|
neuronal guanine nucleotide exchange factor |
| chr5_+_34706936 | 1.44 |
ENSMUST00000179943.3
|
Sh3bp2
|
SH3-domain binding protein 2 |
| chr2_+_81883566 | 1.41 |
ENSMUST00000047527.8
|
Zfp804a
|
zinc finger protein 804A |
| chr5_-_139805661 | 1.41 |
ENSMUST00000147328.2
|
Tmem184a
|
transmembrane protein 184a |
| chr2_+_90677499 | 1.41 |
ENSMUST00000136872.8
ENSMUST00000150232.8 ENSMUST00000111467.4 |
Mtch2
|
mitochondrial carrier 2 |
| chr6_-_52185674 | 1.40 |
ENSMUST00000062829.9
|
Hoxa6
|
homeobox A6 |
| chr8_-_96161466 | 1.39 |
ENSMUST00000213086.2
ENSMUST00000034249.8 |
Cfap20
|
cilia and flagella associated protein 20 |
| chr9_+_44684450 | 1.39 |
ENSMUST00000238800.2
ENSMUST00000147559.8 |
Ift46
|
intraflagellar transport 46 |
| chr11_+_115656246 | 1.39 |
ENSMUST00000093912.11
ENSMUST00000136720.8 ENSMUST00000103034.10 ENSMUST00000141871.8 |
Tmem94
|
transmembrane protein 94 |
| chr2_+_164674801 | 1.39 |
ENSMUST00000103092.9
ENSMUST00000151493.3 ENSMUST00000127650.8 |
Ctsa
|
cathepsin A |
| chr9_-_124075271 | 1.39 |
ENSMUST00000071300.13
ENSMUST00000185949.2 ENSMUST00000177714.8 |
2010315B03Rik
|
RIKEN cDNA 2010315B03 gene |
| chr8_+_12623016 | 1.37 |
ENSMUST00000210276.2
ENSMUST00000010579.8 ENSMUST00000209428.2 |
Spaca7
|
sperm acrosome associated 7 |
| chr2_+_84880776 | 1.36 |
ENSMUST00000111605.9
|
Tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr5_-_38649291 | 1.35 |
ENSMUST00000129099.8
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr1_+_172309337 | 1.34 |
ENSMUST00000127052.8
|
Igsf9
|
immunoglobulin superfamily, member 9 |
| chrX_-_7440480 | 1.34 |
ENSMUST00000115742.9
ENSMUST00000150787.8 |
Ppp1r3f
|
protein phosphatase 1, regulatory subunit 3F |
| chr2_-_71377088 | 1.33 |
ENSMUST00000024159.8
|
Dlx2
|
distal-less homeobox 2 |
| chr5_+_150119860 | 1.33 |
ENSMUST00000202600.4
|
Fry
|
FRY microtubule binding protein |
| chr8_-_70805861 | 1.31 |
ENSMUST00000215817.2
ENSMUST00000075666.8 |
Upf1
|
UPF1 regulator of nonsense transcripts homolog (yeast) |
| chr3_+_102927901 | 1.28 |
ENSMUST00000198180.5
ENSMUST00000197827.5 ENSMUST00000199240.5 ENSMUST00000199420.5 ENSMUST00000199571.5 ENSMUST00000197488.5 |
Csde1
|
cold shock domain containing E1, RNA binding |
| chr5_+_34706911 | 1.22 |
ENSMUST00000118545.8
|
Sh3bp2
|
SH3-domain binding protein 2 |
| chr2_-_155661324 | 1.22 |
ENSMUST00000124586.2
|
BC029722
|
cDNA sequence BC029722 |
| chr9_+_44684324 | 1.20 |
ENSMUST00000214854.3
ENSMUST00000125877.8 |
Ift46
|
intraflagellar transport 46 |
| chr18_+_13139992 | 1.19 |
ENSMUST00000041676.3
ENSMUST00000234084.2 ENSMUST00000234565.2 |
Hrh4
|
histamine receptor H4 |
| chr7_+_141047416 | 1.14 |
ENSMUST00000209988.2
|
Cd151
|
CD151 antigen |
| chr1_-_37904135 | 1.13 |
ENSMUST00000155852.7
ENSMUST00000193669.2 ENSMUST00000041815.15 |
Tsga10
|
testis specific 10 |
| chrX_-_47763355 | 1.11 |
ENSMUST00000053970.4
|
Gpr119
|
G-protein coupled receptor 119 |
| chr1_+_162398084 | 1.11 |
ENSMUST00000132158.8
ENSMUST00000135241.8 |
Vamp4
|
vesicle-associated membrane protein 4 |
| chr7_+_141047298 | 1.11 |
ENSMUST00000106000.10
ENSMUST00000209892.2 ENSMUST00000177840.9 |
Cd151
|
CD151 antigen |
| chr6_-_73198608 | 1.09 |
ENSMUST00000064948.13
ENSMUST00000114040.8 |
Dnah6
|
dynein, axonemal, heavy chain 6 |
| chr2_-_144173615 | 1.07 |
ENSMUST00000103171.10
|
Ovol2
|
ovo like zinc finger 2 |
| chr9_+_65172455 | 1.07 |
ENSMUST00000048762.8
|
Cilp
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| chr7_+_40547608 | 1.06 |
ENSMUST00000044705.12
|
Vstm2b
|
V-set and transmembrane domain containing 2B |
| chr13_-_98951627 | 1.05 |
ENSMUST00000224992.2
ENSMUST00000225840.2 |
Fcho2
|
FCH domain only 2 |
| chr3_-_104419128 | 1.03 |
ENSMUST00000199070.5
ENSMUST00000046316.11 ENSMUST00000198332.2 |
Lrig2
|
leucine-rich repeats and immunoglobulin-like domains 2 |
| chr13_+_43938251 | 1.02 |
ENSMUST00000015540.4
|
Cd83
|
CD83 antigen |
| chr6_-_138635447 | 1.02 |
ENSMUST00000054786.6
|
Igbp1b
|
immunoglobulin (CD79A) binding protein 1b |
| chr18_-_33596468 | 1.02 |
ENSMUST00000171533.9
|
Nrep
|
neuronal regeneration related protein |
| chr4_+_48045143 | 1.01 |
ENSMUST00000030025.10
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr1_-_172034251 | 1.00 |
ENSMUST00000155109.2
|
Pea15a
|
phosphoprotein enriched in astrocytes 15A |
| chr7_+_123061535 | 1.00 |
ENSMUST00000098056.6
|
Aqp8
|
aquaporin 8 |
| chr18_+_70605722 | 0.99 |
ENSMUST00000174118.8
|
Stard6
|
StAR-related lipid transfer (START) domain containing 6 |
| chr19_-_11796085 | 0.97 |
ENSMUST00000211047.2
ENSMUST00000075304.14 ENSMUST00000211641.2 |
Stx3
|
syntaxin 3 |
| chr1_-_162687488 | 0.97 |
ENSMUST00000134098.8
ENSMUST00000111518.3 |
Fmo1
|
flavin containing monooxygenase 1 |
| chr16_-_48814294 | 0.96 |
ENSMUST00000114516.8
|
Dzip3
|
DAZ interacting protein 3, zinc finger |
| chr1_-_132953068 | 0.96 |
ENSMUST00000186617.7
ENSMUST00000067429.10 ENSMUST00000067398.13 ENSMUST00000188090.7 |
Mdm4
|
transformed mouse 3T3 cell double minute 4 |
| chr6_+_17306379 | 0.96 |
ENSMUST00000115455.3
|
Cav1
|
caveolin 1, caveolae protein |
| chr8_+_84874881 | 0.95 |
ENSMUST00000093375.5
|
Brme1
|
break repair meiotic recombinase recruitment factor 1 |
| chr11_+_97689819 | 0.95 |
ENSMUST00000143571.2
|
Lasp1
|
LIM and SH3 protein 1 |
| chr18_+_70605691 | 0.95 |
ENSMUST00000164223.8
|
Stard6
|
StAR-related lipid transfer (START) domain containing 6 |
| chr16_-_48814437 | 0.95 |
ENSMUST00000121869.8
|
Dzip3
|
DAZ interacting protein 3, zinc finger |
| chr7_+_123061497 | 0.92 |
ENSMUST00000033023.10
|
Aqp8
|
aquaporin 8 |
| chr15_+_74828272 | 0.91 |
ENSMUST00000188042.2
|
Ly6e
|
lymphocyte antigen 6 complex, locus E |
| chr3_+_106943472 | 0.90 |
ENSMUST00000052718.5
|
Kcna3
|
potassium voltage-gated channel, shaker-related subfamily, member 3 |
| chr1_+_139349912 | 0.90 |
ENSMUST00000200243.5
ENSMUST00000039867.10 |
Zbtb41
|
zinc finger and BTB domain containing 41 |
| chr2_+_32518402 | 0.90 |
ENSMUST00000156578.8
|
Ak1
|
adenylate kinase 1 |
| chr17_+_34149820 | 0.90 |
ENSMUST00000234226.2
|
Rgl2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr6_+_55429162 | 0.90 |
ENSMUST00000167484.2
|
Adcyap1r1
|
adenylate cyclase activating polypeptide 1 receptor 1 |
| chr3_+_99792699 | 0.89 |
ENSMUST00000164539.2
|
Spag17
|
sperm associated antigen 17 |
| chr19_-_42740898 | 0.87 |
ENSMUST00000237747.2
|
Pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr10_-_80374941 | 0.87 |
ENSMUST00000020383.6
|
Atp8b3
|
ATPase, class I, type 8B, member 3 |
| chr11_-_8989582 | 0.87 |
ENSMUST00000043377.6
|
Sun3
|
Sad1 and UNC84 domain containing 3 |
| chr3_-_6685492 | 0.87 |
ENSMUST00000091364.4
|
1700008P02Rik
|
RIKEN cDNA 1700008P02 gene |
| chr19_+_5497575 | 0.87 |
ENSMUST00000025850.7
ENSMUST00000236774.2 |
Fosl1
|
fos-like antigen 1 |
| chr10_-_33972503 | 0.86 |
ENSMUST00000069125.8
|
Calhm5
|
calcium homeostasis modulator family member 5 |
| chr17_-_27947863 | 0.84 |
ENSMUST00000167489.2
ENSMUST00000138970.3 ENSMUST00000025054.10 ENSMUST00000114870.9 |
Spdef
|
SAM pointed domain containing ets transcription factor |
| chr7_+_27829229 | 0.84 |
ENSMUST00000150948.2
|
9530053A07Rik
|
RIKEN cDNA 9530053A07 gene |
| chr18_-_33597060 | 0.83 |
ENSMUST00000168890.2
|
Nrep
|
neuronal regeneration related protein |
| chr5_-_135378729 | 0.82 |
ENSMUST00000201784.4
ENSMUST00000201791.4 |
Fkbp6
|
FK506 binding protein 6 |
| chr19_-_61129211 | 0.80 |
ENSMUST00000143264.8
ENSMUST00000205854.2 |
Zfp950
|
zinc finger protein 950 |
| chr11_+_98337655 | 0.80 |
ENSMUST00000019456.5
|
Grb7
|
growth factor receptor bound protein 7 |
| chr18_-_64688271 | 0.79 |
ENSMUST00000235459.2
|
Atp8b1
|
ATPase, class I, type 8B, member 1 |
| chr7_+_43959637 | 0.79 |
ENSMUST00000107938.8
|
Shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr8_-_68270936 | 0.78 |
ENSMUST00000120071.8
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr17_+_9207165 | 0.78 |
ENSMUST00000024650.12
|
1700010I14Rik
|
RIKEN cDNA 1700010I14 gene |
| chr17_+_31514780 | 0.77 |
ENSMUST00000237460.2
|
Slc37a1
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 1 |
| chr1_-_182110303 | 0.77 |
ENSMUST00000035295.6
|
Degs1
|
delta(4)-desaturase, sphingolipid 1 |
| chr8_-_69848167 | 0.75 |
ENSMUST00000072427.7
ENSMUST00000213012.2 ENSMUST00000239456.2 |
Gm10033
|
predicted gene 10033 |
| chr18_+_90597896 | 0.75 |
ENSMUST00000211710.2
ENSMUST00000209969.3 |
Gm45871
|
predicted gene 45871 |
| chr11_+_116547932 | 0.75 |
ENSMUST00000116318.3
|
Prcd
|
photoreceptor disc component |
| chr16_+_87151073 | 0.74 |
ENSMUST00000054442.11
ENSMUST00000118310.8 ENSMUST00000120284.8 ENSMUST00000118115.2 |
N6amt1
|
N-6 adenine-specific DNA methyltransferase 1 (putative) |
| chr13_+_83672389 | 0.74 |
ENSMUST00000200394.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr6_+_83055581 | 0.72 |
ENSMUST00000177177.8
ENSMUST00000176089.2 |
Pcgf1
|
polycomb group ring finger 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 33.1 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 5.4 | 21.5 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 3.3 | 13.0 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 2.9 | 11.5 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 1.9 | 7.7 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 1.4 | 5.8 | GO:0045963 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 1.3 | 8.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) positive regulation of phospholipase A2 activity(GO:0032430) |
| 1.3 | 5.1 | GO:0032429 | regulation of phospholipase A2 activity(GO:0032429) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 1.1 | 3.2 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 1.1 | 3.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 1.0 | 2.9 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.8 | 2.5 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.8 | 3.2 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.8 | 2.4 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.7 | 8.9 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.7 | 24.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) response to stilbenoid(GO:0035634) |
| 0.7 | 4.1 | GO:0032824 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.7 | 5.4 | GO:0043651 | linoleic acid metabolic process(GO:0043651) positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.7 | 2.0 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.6 | 10.4 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.6 | 2.6 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.6 | 3.8 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.6 | 14.3 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.6 | 1.7 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.6 | 2.8 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.5 | 59.3 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.5 | 6.3 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.5 | 1.5 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.5 | 15.0 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.5 | 5.3 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.5 | 5.5 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.4 | 3.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.4 | 1.3 | GO:1902871 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.4 | 2.5 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.4 | 2.3 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.4 | 2.2 | GO:0060336 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.4 | 2.8 | GO:0009313 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.3 | 3.4 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.3 | 2.3 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.3 | 1.3 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.3 | 4.7 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.3 | 8.0 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.3 | 10.6 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.3 | 4.7 | GO:0042407 | cristae formation(GO:0042407) |
| 0.3 | 1.9 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.3 | 1.3 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.3 | 1.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.3 | 1.0 | GO:1900623 | positive regulation of mast cell cytokine production(GO:0032765) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.2 | 1.4 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.2 | 4.0 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 0.9 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.2 | 0.6 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.2 | 0.4 | GO:0042374 | menaquinone metabolic process(GO:0009233) phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.2 | 1.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 3.0 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.2 | 0.5 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.2 | 1.7 | GO:0098881 | synaptic vesicle docking(GO:0016081) exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.2 | 1.6 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.2 | 28.0 | GO:0009308 | amine metabolic process(GO:0009308) |
| 0.2 | 0.9 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.1 | 2.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 0.9 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 1.4 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.1 | 1.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 1.7 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 1.1 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.1 | 0.6 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.1 | 0.3 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.1 | 1.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.7 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.1 | 0.8 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.1 | 2.6 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.1 | 0.3 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.1 | 1.4 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.4 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.1 | 1.0 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.1 | 1.1 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) regulation of establishment of protein localization to plasma membrane(GO:0090003) |
| 0.1 | 1.5 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 0.9 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 1.3 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.1 | 0.3 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.8 | GO:0046959 | habituation(GO:0046959) |
| 0.1 | 0.3 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 1.5 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 0.3 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 | 0.9 | GO:0048241 | epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) |
| 0.1 | 2.2 | GO:2001273 | regulation of glucose import in response to insulin stimulus(GO:2001273) |
| 0.1 | 1.8 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 0.2 | GO:2000506 | positive regulation of small intestine smooth muscle contraction(GO:1904349) negative regulation of energy homeostasis(GO:2000506) |
| 0.1 | 1.3 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.4 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.1 | 0.7 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.5 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 0.3 | GO:0050968 | sensory perception of sour taste(GO:0050915) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.1 | 0.9 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 2.7 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 1.0 | GO:0003181 | atrioventricular valve morphogenesis(GO:0003181) |
| 0.1 | 0.4 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.1 | 1.6 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 1.3 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.6 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 1.9 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.6 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 7.8 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.0 | 2.9 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) |
| 0.0 | 3.5 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 2.6 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.7 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.0 | 1.4 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 1.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.8 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.8 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 5.2 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 0.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.5 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.3 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 5.9 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 1.2 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.4 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.7 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 1.5 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 1.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 1.1 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 2.7 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.2 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 0.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 3.7 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.4 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 1.9 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.7 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.0 | 0.0 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 | 0.3 | GO:0048485 | sympathetic nervous system development(GO:0048485) |
| 0.0 | 0.2 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.0 | 0.3 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.5 | GO:0043030 | regulation of macrophage activation(GO:0043030) |
| 0.0 | 1.7 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.3 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.7 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.0 | 0.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 1.2 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.0 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 2.2 | 33.1 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 1.5 | 6.0 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.6 | 4.7 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.5 | 8.9 | GO:0043203 | axon hillock(GO:0043203) |
| 0.5 | 1.9 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.4 | 1.5 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.3 | 24.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 1.7 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.2 | 1.9 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.2 | 4.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 0.9 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.2 | 1.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 58.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.2 | 2.6 | GO:0000243 | commitment complex(GO:0000243) |
| 0.2 | 1.0 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 2.6 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 5.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 2.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 3.8 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 8.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 1.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 0.6 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 1.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.9 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 10.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 2.6 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.9 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 2.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 13.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 2.4 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 2.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.5 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 4.9 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 1.1 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.7 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 1.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 3.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 1.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 2.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 4.5 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.4 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 5.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 5.6 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 1.4 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 39.2 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 2.3 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.0 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 1.3 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 3.5 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 6.2 | GO:0019866 | organelle inner membrane(GO:0019866) |
| 0.0 | 0.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 3.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.2 | 57.6 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 3.6 | 21.5 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 3.5 | 28.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 2.4 | 7.1 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 2.2 | 13.0 | GO:0032810 | sterol response element binding(GO:0032810) |
| 1.9 | 7.7 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 1.9 | 5.8 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 1.7 | 8.4 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 1.3 | 33.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 1.1 | 3.4 | GO:0008124 | 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 1.1 | 4.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 1.0 | 3.8 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.9 | 10.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.8 | 12.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.8 | 24.3 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.7 | 3.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.7 | 4.1 | GO:0016019 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.6 | 2.8 | GO:0052794 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.5 | 6.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.5 | 10.4 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.5 | 2.4 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.5 | 1.8 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.4 | 2.5 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.4 | 2.3 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.4 | 3.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.3 | 1.7 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.3 | 5.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.3 | 4.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.3 | 2.6 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.3 | 11.1 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.3 | 2.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.3 | 5.3 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) aromatase activity(GO:0070330) |
| 0.3 | 1.7 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.3 | 1.9 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.3 | 1.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.3 | 6.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.3 | 2.7 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.2 | 2.9 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 4.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 1.6 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.2 | 0.8 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.2 | 1.7 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.2 | 1.1 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.2 | 5.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 3.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.2 | 2.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.2 | 1.4 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 2.6 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 9.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 5.6 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 3.2 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.4 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 2.3 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 7.7 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 1.9 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 1.9 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 1.4 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 10.6 | GO:0020037 | heme binding(GO:0020037) |
| 0.1 | 2.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 8.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 0.9 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 2.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.8 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.2 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 0.1 | 1.5 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.1 | 1.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.9 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.4 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 2.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 2.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 1.8 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.0 | 0.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 2.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 6.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.9 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 1.5 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.6 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 1.1 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 4.4 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 1.3 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 1.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 2.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.3 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 1.0 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 1.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.3 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.4 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.5 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.8 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.3 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.3 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 32.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.3 | 15.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 9.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 2.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 2.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 11.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.9 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 2.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 2.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 2.7 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 5.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.6 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.2 | 21.5 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 1.1 | 12.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.9 | 12.0 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.7 | 10.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.3 | 2.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 6.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 31.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 4.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 12.0 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.2 | 2.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 9.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 1.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 14.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 4.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 1.7 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.9 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 2.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 2.9 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 1.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.9 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 3.0 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 3.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 2.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 7.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 6.5 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |