Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
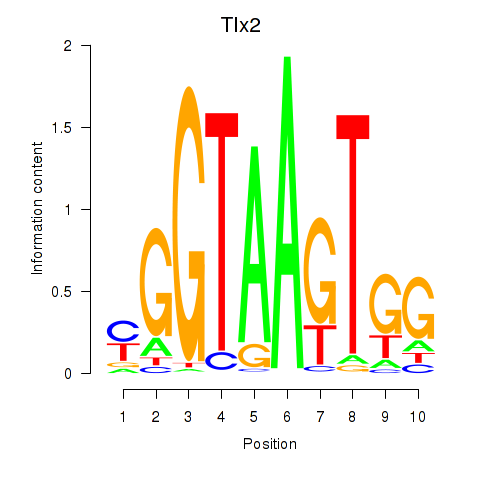
Results for Tlx2
Z-value: 1.56
Transcription factors associated with Tlx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tlx2
|
ENSMUSG00000068327.6 | Tlx2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tlx2 | mm39_v1_chr6_-_83047206_83047361 | 0.28 | 1.0e-01 | Click! |
Activity profile of Tlx2 motif
Sorted Z-values of Tlx2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tlx2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_90576285 | 10.30 |
ENSMUST00000069927.10
|
S100a8
|
S100 calcium binding protein A8 (calgranulin A) |
| chr3_-_90603013 | 9.81 |
ENSMUST00000069960.12
ENSMUST00000117167.2 |
S100a9
|
S100 calcium binding protein A9 (calgranulin B) |
| chr10_+_79722081 | 7.92 |
ENSMUST00000046091.7
|
Elane
|
elastase, neutrophil expressed |
| chr14_+_46997984 | 7.31 |
ENSMUST00000067426.6
|
Cdkn3
|
cyclin-dependent kinase inhibitor 3 |
| chr8_+_81220410 | 7.05 |
ENSMUST00000063359.8
|
Gypa
|
glycophorin A |
| chr10_+_79715448 | 6.12 |
ENSMUST00000006679.15
|
Prtn3
|
proteinase 3 |
| chr8_+_84682136 | 5.78 |
ENSMUST00000005607.9
|
Asf1b
|
anti-silencing function 1B histone chaperone |
| chr3_+_14951478 | 5.66 |
ENSMUST00000029078.9
|
Car2
|
carbonic anhydrase 2 |
| chr12_+_24758968 | 5.50 |
ENSMUST00000154588.2
|
Rrm2
|
ribonucleotide reductase M2 |
| chr10_+_43455157 | 5.22 |
ENSMUST00000058714.10
|
Cd24a
|
CD24a antigen |
| chr3_+_14951264 | 5.21 |
ENSMUST00000192609.6
|
Car2
|
carbonic anhydrase 2 |
| chr16_+_48692976 | 5.14 |
ENSMUST00000065666.6
|
Retnlg
|
resistin like gamma |
| chr6_-_41681273 | 4.62 |
ENSMUST00000031899.14
|
Kel
|
Kell blood group |
| chr3_-_84386724 | 4.62 |
ENSMUST00000091002.8
|
Fhdc1
|
FH2 domain containing 1 |
| chr12_+_24758240 | 4.53 |
ENSMUST00000020980.12
|
Rrm2
|
ribonucleotide reductase M2 |
| chr3_-_98247237 | 4.52 |
ENSMUST00000065793.12
|
Phgdh
|
3-phosphoglycerate dehydrogenase |
| chr2_-_150510116 | 4.35 |
ENSMUST00000028944.4
|
Acss1
|
acyl-CoA synthetase short-chain family member 1 |
| chrX_-_92875712 | 4.26 |
ENSMUST00000045748.7
|
Pdk3
|
pyruvate dehydrogenase kinase, isoenzyme 3 |
| chr19_-_46033353 | 4.25 |
ENSMUST00000026252.14
ENSMUST00000156585.9 ENSMUST00000185355.7 ENSMUST00000152946.8 |
Ldb1
|
LIM domain binding 1 |
| chr12_+_24758724 | 4.16 |
ENSMUST00000153058.8
|
Rrm2
|
ribonucleotide reductase M2 |
| chr14_+_46998004 | 3.98 |
ENSMUST00000227149.2
|
Cdkn3
|
cyclin-dependent kinase inhibitor 3 |
| chr18_-_57058581 | 3.95 |
ENSMUST00000102912.8
|
Marchf3
|
membrane associated ring-CH-type finger 3 |
| chr7_-_126736979 | 3.93 |
ENSMUST00000049931.6
|
Spn
|
sialophorin |
| chr11_+_115790768 | 3.70 |
ENSMUST00000152171.8
|
Smim5
|
small integral membrane protein 5 |
| chr11_-_11987391 | 3.69 |
ENSMUST00000093321.12
|
Grb10
|
growth factor receptor bound protein 10 |
| chrX_+_99669343 | 3.45 |
ENSMUST00000048962.4
|
Kif4
|
kinesin family member 4 |
| chrX_+_8137881 | 3.36 |
ENSMUST00000115590.2
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr19_-_5776268 | 3.25 |
ENSMUST00000075606.6
ENSMUST00000236215.2 ENSMUST00000235730.2 ENSMUST00000237081.2 ENSMUST00000049295.15 |
Ehbp1l1
|
EH domain binding protein 1-like 1 |
| chr1_+_172327569 | 3.25 |
ENSMUST00000111230.8
|
Tagln2
|
transgelin 2 |
| chr7_+_78563964 | 3.14 |
ENSMUST00000120331.4
|
Isg20
|
interferon-stimulated protein |
| chr3_-_96201248 | 3.00 |
ENSMUST00000029748.8
|
Fcgr1
|
Fc receptor, IgG, high affinity I |
| chr4_+_130253925 | 2.97 |
ENSMUST00000105994.4
|
Snrnp40
|
small nuclear ribonucleoprotein 40 (U5) |
| chr15_+_103148824 | 2.94 |
ENSMUST00000036004.16
ENSMUST00000087351.9 ENSMUST00000231141.2 |
Hnrnpa1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr8_+_73488496 | 2.92 |
ENSMUST00000058099.9
|
F2rl3
|
coagulation factor II (thrombin) receptor-like 3 |
| chr4_-_154110324 | 2.88 |
ENSMUST00000130175.8
ENSMUST00000182151.8 |
Smim1
|
small integral membrane protein 1 |
| chr1_-_75482975 | 2.84 |
ENSMUST00000113567.10
ENSMUST00000113565.3 |
Obsl1
|
obscurin-like 1 |
| chr11_+_68936457 | 2.78 |
ENSMUST00000108666.8
ENSMUST00000021277.6 |
Aurkb
|
aurora kinase B |
| chr4_-_154110383 | 2.74 |
ENSMUST00000132541.8
ENSMUST00000143047.8 |
Smim1
|
small integral membrane protein 1 |
| chr2_+_84818538 | 2.72 |
ENSMUST00000028466.12
|
Prg3
|
proteoglycan 3 |
| chr4_-_133600308 | 2.71 |
ENSMUST00000137486.3
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr1_+_172327812 | 2.71 |
ENSMUST00000192460.2
|
Tagln2
|
transgelin 2 |
| chr11_+_115790951 | 2.69 |
ENSMUST00000142089.2
ENSMUST00000131566.2 |
Smim5
|
small integral membrane protein 5 |
| chr11_-_75918551 | 2.69 |
ENSMUST00000021207.7
|
Rflnb
|
refilin B |
| chr10_-_62178453 | 2.65 |
ENSMUST00000143179.2
ENSMUST00000130422.8 |
Hk1
|
hexokinase 1 |
| chrX_-_36253309 | 2.62 |
ENSMUST00000060474.14
ENSMUST00000053456.11 ENSMUST00000115239.10 |
Septin6
|
septin 6 |
| chr7_-_44181477 | 2.61 |
ENSMUST00000098483.9
ENSMUST00000035323.6 |
Spib
|
Spi-B transcription factor (Spi-1/PU.1 related) |
| chr8_+_71047110 | 2.58 |
ENSMUST00000019283.10
ENSMUST00000210005.2 |
Isyna1
|
myo-inositol 1-phosphate synthase A1 |
| chr17_+_18108086 | 2.52 |
ENSMUST00000149944.2
|
Fpr2
|
formyl peptide receptor 2 |
| chr2_-_129139125 | 2.46 |
ENSMUST00000052708.7
|
Ckap2l
|
cytoskeleton associated protein 2-like |
| chr5_+_45827249 | 2.46 |
ENSMUST00000117396.3
|
Ncapg
|
non-SMC condensin I complex, subunit G |
| chr7_+_127503812 | 2.45 |
ENSMUST00000151451.3
ENSMUST00000124533.3 ENSMUST00000206745.2 ENSMUST00000206140.2 |
Bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr7_+_78564062 | 2.39 |
ENSMUST00000205981.2
|
Isg20
|
interferon-stimulated protein |
| chr16_+_49675682 | 2.37 |
ENSMUST00000114496.3
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr8_-_71725011 | 2.36 |
ENSMUST00000110071.3
|
Haus8
|
4HAUS augmin-like complex, subunit 8 |
| chr11_+_83328503 | 2.35 |
ENSMUST00000037378.6
|
1700020L24Rik
|
RIKEN cDNA 1700020L24 gene |
| chr7_-_126399574 | 2.31 |
ENSMUST00000106348.8
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr17_+_18108102 | 2.30 |
ENSMUST00000054871.12
ENSMUST00000064068.5 |
Fpr3
Fpr2
|
formyl peptide receptor 3 formyl peptide receptor 2 |
| chr8_-_123278054 | 2.26 |
ENSMUST00000156333.9
ENSMUST00000067252.14 |
Piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr7_+_30193047 | 2.24 |
ENSMUST00000058280.13
ENSMUST00000133318.8 ENSMUST00000142575.8 ENSMUST00000131040.2 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chr1_+_134890288 | 2.23 |
ENSMUST00000027687.8
|
Ube2t
|
ubiquitin-conjugating enzyme E2T |
| chr11_+_106642052 | 2.18 |
ENSMUST00000147326.9
ENSMUST00000182896.8 ENSMUST00000182908.8 ENSMUST00000086353.11 |
Milr1
|
mast cell immunoglobulin like receptor 1 |
| chr16_-_36154692 | 2.15 |
ENSMUST00000114850.3
|
Cstdc6
|
cystatin domain containing 6 |
| chr7_-_126399208 | 2.08 |
ENSMUST00000133514.8
ENSMUST00000151137.8 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr7_-_4815542 | 2.07 |
ENSMUST00000079496.9
|
Ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr8_-_106660470 | 2.07 |
ENSMUST00000034368.8
|
Ctrl
|
chymotrypsin-like |
| chr7_-_126399778 | 2.03 |
ENSMUST00000141355.4
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr7_+_127503251 | 1.93 |
ENSMUST00000071056.14
|
Bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr10_+_79824418 | 1.92 |
ENSMUST00000004784.11
ENSMUST00000105374.2 |
Cnn2
|
calponin 2 |
| chr16_+_49676130 | 1.90 |
ENSMUST00000230641.2
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr16_-_36486429 | 1.90 |
ENSMUST00000089620.11
|
Cd86
|
CD86 antigen |
| chr11_+_121312212 | 1.87 |
ENSMUST00000038096.8
|
Fn3krp
|
fructosamine 3 kinase related protein |
| chr8_-_124675939 | 1.84 |
ENSMUST00000044795.8
|
Nup133
|
nucleoporin 133 |
| chr13_-_49806231 | 1.84 |
ENSMUST00000021818.9
|
Cenpp
|
centromere protein P |
| chr19_-_7218363 | 1.81 |
ENSMUST00000236769.2
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr10_-_62343516 | 1.80 |
ENSMUST00000020271.13
|
Srgn
|
serglycin |
| chr8_-_25592385 | 1.78 |
ENSMUST00000064883.14
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr3_-_88410495 | 1.76 |
ENSMUST00000120377.8
ENSMUST00000029699.13 |
Lmna
|
lamin A |
| chr19_-_7218512 | 1.75 |
ENSMUST00000025675.11
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr16_+_49675969 | 1.73 |
ENSMUST00000229101.2
ENSMUST00000230836.2 ENSMUST00000229206.2 ENSMUST00000084838.14 ENSMUST00000230281.2 |
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr6_-_56878854 | 1.73 |
ENSMUST00000101367.9
|
Nt5c3
|
5'-nucleotidase, cytosolic III |
| chr6_+_83003253 | 1.69 |
ENSMUST00000204891.2
|
M1ap
|
meiosis 1 associated protein |
| chr19_+_8734437 | 1.69 |
ENSMUST00000010241.14
|
Nxf1
|
nuclear RNA export factor 1 |
| chr1_+_34511793 | 1.68 |
ENSMUST00000188972.3
|
Ptpn18
|
protein tyrosine phosphatase, non-receptor type 18 |
| chr5_-_67973195 | 1.67 |
ENSMUST00000141443.2
|
Atp8a1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 |
| chr5_-_5799315 | 1.67 |
ENSMUST00000015796.9
|
Steap1
|
six transmembrane epithelial antigen of the prostate 1 |
| chr7_-_16657825 | 1.67 |
ENSMUST00000019514.10
|
Calm3
|
calmodulin 3 |
| chr11_-_106051533 | 1.67 |
ENSMUST00000106875.2
|
Limd2
|
LIM domain containing 2 |
| chr2_-_119985078 | 1.66 |
ENSMUST00000028755.8
|
Ehd4
|
EH-domain containing 4 |
| chr5_+_108048697 | 1.66 |
ENSMUST00000153590.2
|
Rpl5
|
ribosomal protein L5 |
| chr11_-_61384998 | 1.65 |
ENSMUST00000101085.9
ENSMUST00000079080.13 ENSMUST00000108714.2 |
Mapk7
|
mitogen-activated protein kinase 7 |
| chr5_+_108048367 | 1.63 |
ENSMUST00000082223.13
|
Rpl5
|
ribosomal protein L5 |
| chr7_-_45084012 | 1.63 |
ENSMUST00000107771.12
ENSMUST00000211666.2 |
Ruvbl2
|
RuvB-like protein 2 |
| chr2_+_129040677 | 1.63 |
ENSMUST00000028880.10
|
Slc20a1
|
solute carrier family 20, member 1 |
| chr12_-_113552322 | 1.63 |
ENSMUST00000103443.3
|
Ighv2-2
|
immunoglobulin heavy variable 2-2 |
| chr7_-_115933367 | 1.61 |
ENSMUST00000205490.2
ENSMUST00000170953.3 |
Rps13
|
ribosomal protein S13 |
| chr6_+_35229628 | 1.60 |
ENSMUST00000130875.8
|
1810058I24Rik
|
RIKEN cDNA 1810058I24 gene |
| chr11_-_86999481 | 1.57 |
ENSMUST00000051395.9
|
Prr11
|
proline rich 11 |
| chr3_+_68912043 | 1.57 |
ENSMUST00000042901.15
|
Smc4
|
structural maintenance of chromosomes 4 |
| chr8_-_25591737 | 1.56 |
ENSMUST00000098866.11
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr19_+_8734463 | 1.56 |
ENSMUST00000184970.9
|
Nxf1
|
nuclear RNA export factor 1 |
| chr7_+_28136861 | 1.56 |
ENSMUST00000108292.9
ENSMUST00000108289.8 |
Gmfg
|
glia maturation factor, gamma |
| chr4_+_129941633 | 1.53 |
ENSMUST00000044565.15
ENSMUST00000132251.2 |
Col16a1
|
collagen, type XVI, alpha 1 |
| chr5_+_31409021 | 1.51 |
ENSMUST00000054829.13
ENSMUST00000201625.4 ENSMUST00000201937.4 |
Krtcap3
|
keratinocyte associated protein 3 |
| chr6_+_38895902 | 1.51 |
ENSMUST00000003017.13
|
Tbxas1
|
thromboxane A synthase 1, platelet |
| chr1_+_34498836 | 1.50 |
ENSMUST00000027302.14
ENSMUST00000190122.2 |
Ptpn18
|
protein tyrosine phosphatase, non-receptor type 18 |
| chr10_+_79852750 | 1.49 |
ENSMUST00000105373.8
|
Arhgap45
|
Rho GTPase activating protein 45 |
| chr7_+_3337591 | 1.49 |
ENSMUST00000203328.4
|
Myadm
|
myeloid-associated differentiation marker |
| chr6_+_35229589 | 1.48 |
ENSMUST00000152147.8
|
1810058I24Rik
|
RIKEN cDNA 1810058I24 gene |
| chr10_+_79852487 | 1.46 |
ENSMUST00000099501.10
|
Arhgap45
|
Rho GTPase activating protein 45 |
| chr17_-_43813664 | 1.46 |
ENSMUST00000024707.9
ENSMUST00000117137.8 |
Mep1a
|
meprin 1 alpha |
| chr1_-_45964730 | 1.44 |
ENSMUST00000027137.11
|
Slc40a1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr17_+_35413415 | 1.43 |
ENSMUST00000025262.6
ENSMUST00000173600.2 |
Ltb
|
lymphotoxin B |
| chr3_+_159545309 | 1.41 |
ENSMUST00000068952.10
ENSMUST00000198878.2 |
Wls
|
wntless WNT ligand secretion mediator |
| chr19_+_8734503 | 1.41 |
ENSMUST00000183939.2
|
Nxf1
|
nuclear RNA export factor 1 |
| chr5_-_38719025 | 1.39 |
ENSMUST00000005234.13
|
Wdr1
|
WD repeat domain 1 |
| chr11_+_63019799 | 1.37 |
ENSMUST00000108702.8
|
Pmp22
|
peripheral myelin protein 22 |
| chr8_-_8740471 | 1.36 |
ENSMUST00000048545.10
|
Arglu1
|
arginine and glutamate rich 1 |
| chr7_+_43057611 | 1.35 |
ENSMUST00000005592.7
|
Siglecg
|
sialic acid binding Ig-like lectin G |
| chr15_-_78413780 | 1.35 |
ENSMUST00000229185.2
|
C1qtnf6
|
C1q and tumor necrosis factor related protein 6 |
| chr15_-_34443355 | 1.35 |
ENSMUST00000009039.6
|
Rpl30
|
ribosomal protein L30 |
| chr13_-_59917569 | 1.34 |
ENSMUST00000057115.7
|
Isca1
|
iron-sulfur cluster assembly 1 |
| chr5_-_22755274 | 1.34 |
ENSMUST00000030872.12
|
Orc5
|
origin recognition complex, subunit 5 |
| chr2_+_127967951 | 1.34 |
ENSMUST00000089634.12
ENSMUST00000019281.14 ENSMUST00000110341.9 ENSMUST00000103211.8 ENSMUST00000103210.2 |
Bcl2l11
|
BCL2-like 11 (apoptosis facilitator) |
| chr19_-_4240984 | 1.34 |
ENSMUST00000045864.4
|
Tbc1d10c
|
TBC1 domain family, member 10c |
| chr6_-_115785695 | 1.33 |
ENSMUST00000081840.6
|
Rpl32
|
ribosomal protein L32 |
| chr19_+_58748132 | 1.33 |
ENSMUST00000026081.5
|
Pnliprp2
|
pancreatic lipase-related protein 2 |
| chr4_-_46138397 | 1.33 |
ENSMUST00000144495.2
ENSMUST00000107770.2 ENSMUST00000156021.2 ENSMUST00000107772.8 |
Tstd2
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 2 |
| chr13_-_12272962 | 1.32 |
ENSMUST00000099856.6
|
Mtr
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr11_-_78642480 | 1.31 |
ENSMUST00000059468.6
|
Ccnq
|
cyclin Q |
| chr14_+_32043944 | 1.30 |
ENSMUST00000022480.8
ENSMUST00000228529.2 |
Ogdhl
|
oxoglutarate dehydrogenase-like |
| chr7_-_19093383 | 1.27 |
ENSMUST00000047036.10
|
Cd3eap
|
CD3E antigen, epsilon polypeptide associated protein |
| chr6_+_38918327 | 1.26 |
ENSMUST00000160963.2
|
Tbxas1
|
thromboxane A synthase 1, platelet |
| chr19_-_4251589 | 1.26 |
ENSMUST00000237923.2
ENSMUST00000025740.8 ENSMUST00000237723.2 |
Rad9a
|
RAD9 checkpoint clamp component A |
| chr7_+_46496929 | 1.26 |
ENSMUST00000132157.2
ENSMUST00000210631.2 |
Ldha
|
lactate dehydrogenase A |
| chr15_-_34443786 | 1.26 |
ENSMUST00000079735.12
|
Rpl30
|
ribosomal protein L30 |
| chr5_-_137834470 | 1.25 |
ENSMUST00000110980.2
ENSMUST00000058897.11 ENSMUST00000199028.2 |
Pilra
|
paired immunoglobin-like type 2 receptor alpha |
| chr6_+_83156550 | 1.24 |
ENSMUST00000113919.10
ENSMUST00000113918.8 ENSMUST00000141680.8 |
Dctn1
|
dynactin 1 |
| chr11_+_101333238 | 1.23 |
ENSMUST00000107249.8
|
Rpl27
|
ribosomal protein L27 |
| chr12_+_17398421 | 1.23 |
ENSMUST00000046011.12
|
Nol10
|
nucleolar protein 10 |
| chr12_-_111638722 | 1.22 |
ENSMUST00000001304.9
|
Ckb
|
creatine kinase, brain |
| chr1_-_36312482 | 1.22 |
ENSMUST00000056946.8
|
Neurl3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr3_-_59038634 | 1.21 |
ENSMUST00000200358.2
ENSMUST00000197220.2 |
P2ry14
|
purinergic receptor P2Y, G-protein coupled, 14 |
| chr1_-_91386976 | 1.20 |
ENSMUST00000069620.10
|
Per2
|
period circadian clock 2 |
| chr18_+_75133519 | 1.20 |
ENSMUST00000079716.6
|
Rpl17
|
ribosomal protein L17 |
| chrX_+_74557905 | 1.19 |
ENSMUST00000114070.10
ENSMUST00000033540.6 |
Vbp1
|
von Hippel-Lindau binding protein 1 |
| chr3_-_59038031 | 1.18 |
ENSMUST00000091112.6
ENSMUST00000065220.13 |
P2ry14
|
purinergic receptor P2Y, G-protein coupled, 14 |
| chr3_+_103078971 | 1.18 |
ENSMUST00000005830.15
|
Bcas2
|
breast carcinoma amplified sequence 2 |
| chr4_+_140428777 | 1.17 |
ENSMUST00000138808.8
ENSMUST00000038893.6 |
Rcc2
|
regulator of chromosome condensation 2 |
| chr5_-_137145030 | 1.17 |
ENSMUST00000054384.6
ENSMUST00000152207.2 |
Trim56
|
tripartite motif-containing 56 |
| chr15_-_34443054 | 1.16 |
ENSMUST00000142643.2
|
Rpl30
|
ribosomal protein L30 |
| chr4_+_43506966 | 1.16 |
ENSMUST00000030183.10
|
Car9
|
carbonic anhydrase 9 |
| chr11_+_106642079 | 1.15 |
ENSMUST00000183111.8
ENSMUST00000106794.9 |
Milr1
|
mast cell immunoglobulin like receptor 1 |
| chr16_+_13981139 | 1.14 |
ENSMUST00000023359.13
ENSMUST00000117958.8 |
Nde1
|
nudE neurodevelopment protein 1 |
| chr10_-_128755127 | 1.12 |
ENSMUST00000149961.2
ENSMUST00000026406.14 |
Rdh5
|
retinol dehydrogenase 5 |
| chr13_-_76166789 | 1.12 |
ENSMUST00000179078.9
ENSMUST00000167271.9 |
Rfesd
|
Rieske (Fe-S) domain containing |
| chr16_+_22710785 | 1.12 |
ENSMUST00000023583.7
ENSMUST00000232098.2 |
Ahsg
|
alpha-2-HS-glycoprotein |
| chr6_-_31540913 | 1.11 |
ENSMUST00000026698.8
|
Podxl
|
podocalyxin-like |
| chr2_-_181335518 | 1.10 |
ENSMUST00000108776.8
ENSMUST00000108771.2 |
Rgs19
|
regulator of G-protein signaling 19 |
| chr16_+_13981180 | 1.10 |
ENSMUST00000115795.9
|
Nde1
|
nudE neurodevelopment protein 1 |
| chr19_-_5779648 | 1.09 |
ENSMUST00000116558.3
ENSMUST00000099955.4 ENSMUST00000161368.2 |
Fam89b
|
family with sequence similarity 89, member B |
| chr4_-_41275091 | 1.08 |
ENSMUST00000030143.13
ENSMUST00000108068.8 |
Ubap2
|
ubiquitin-associated protein 2 |
| chr7_+_109118345 | 1.07 |
ENSMUST00000143107.2
|
Rpl27a
|
ribosomal protein L27A |
| chr14_-_54891073 | 1.07 |
ENSMUST00000126166.8
ENSMUST00000141453.8 ENSMUST00000150371.8 ENSMUST00000123875.2 ENSMUST00000022794.14 ENSMUST00000148754.10 |
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr19_-_4241034 | 1.07 |
ENSMUST00000237495.2
|
Tbc1d10c
|
TBC1 domain family, member 10c |
| chr15_-_103148239 | 1.05 |
ENSMUST00000118152.8
|
Cbx5
|
chromobox 5 |
| chr5_-_116162415 | 1.05 |
ENSMUST00000031486.14
ENSMUST00000111999.8 |
Prkab1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr3_-_129834788 | 1.05 |
ENSMUST00000168644.3
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae) |
| chr9_+_107468146 | 1.05 |
ENSMUST00000195746.2
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr5_-_137856280 | 1.05 |
ENSMUST00000110978.7
ENSMUST00000199387.2 ENSMUST00000196195.2 |
Pilrb1
|
paired immunoglobin-like type 2 receptor beta 1 |
| chr3_+_103767581 | 1.04 |
ENSMUST00000029433.9
|
Ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr11_+_101333115 | 1.03 |
ENSMUST00000077856.13
|
Rpl27
|
ribosomal protein L27 |
| chr19_+_46385321 | 1.01 |
ENSMUST00000039922.13
ENSMUST00000111867.9 ENSMUST00000120778.8 |
Sufu
|
SUFU negative regulator of hedgehog signaling |
| chr4_-_59783780 | 1.01 |
ENSMUST00000107526.8
ENSMUST00000095063.11 |
Inip
|
INTS3 and NABP interacting protein |
| chr7_-_45083688 | 0.97 |
ENSMUST00000210439.2
|
Ruvbl2
|
RuvB-like protein 2 |
| chr18_+_36661198 | 0.97 |
ENSMUST00000237174.2
ENSMUST00000236124.2 ENSMUST00000236779.2 ENSMUST00000235181.2 ENSMUST00000074298.13 ENSMUST00000115694.3 ENSMUST00000236126.2 |
Slc4a9
|
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
| chr5_-_105198913 | 0.97 |
ENSMUST00000112718.5
|
Gbp8
|
guanylate-binding protein 8 |
| chr7_-_131012202 | 0.97 |
ENSMUST00000207243.2
ENSMUST00000128432.3 ENSMUST00000121033.8 ENSMUST00000046306.15 |
Ikzf5
|
IKAROS family zinc finger 5 |
| chr19_+_6451667 | 0.97 |
ENSMUST00000113471.3
ENSMUST00000113469.3 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr4_-_129452148 | 0.96 |
ENSMUST00000167288.8
ENSMUST00000134336.3 |
Lck
|
lymphocyte protein tyrosine kinase |
| chr4_-_135780660 | 0.96 |
ENSMUST00000102536.11
|
Rpl11
|
ribosomal protein L11 |
| chr11_+_95728042 | 0.95 |
ENSMUST00000107712.8
|
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chrX_-_100310959 | 0.95 |
ENSMUST00000135038.2
|
Il2rg
|
interleukin 2 receptor, gamma chain |
| chr4_-_129452180 | 0.95 |
ENSMUST00000067240.11
|
Lck
|
lymphocyte protein tyrosine kinase |
| chr9_+_119274009 | 0.94 |
ENSMUST00000035094.14
|
Exog
|
endo/exonuclease (5'-3'), endonuclease G-like |
| chr16_+_17437205 | 0.94 |
ENSMUST00000006053.13
ENSMUST00000231311.2 ENSMUST00000163476.10 ENSMUST00000231257.2 ENSMUST00000165363.10 ENSMUST00000232043.2 ENSMUST00000090159.13 ENSMUST00000232442.2 |
Smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr2_+_112209548 | 0.93 |
ENSMUST00000028552.4
|
Katnbl1
|
katanin p80 subunit B like 1 |
| chr5_-_86213439 | 0.92 |
ENSMUST00000031170.10
|
Cenpc1
|
centromere protein C1 |
| chr1_+_74545203 | 0.91 |
ENSMUST00000087215.7
|
Cnot9
|
CCR4-NOT transcription complex, subunit 9 |
| chr15_-_64184485 | 0.90 |
ENSMUST00000177083.8
ENSMUST00000177371.8 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr3_+_94320548 | 0.90 |
ENSMUST00000166032.8
ENSMUST00000200486.5 ENSMUST00000196386.5 ENSMUST00000045245.10 ENSMUST00000197901.5 ENSMUST00000198041.2 |
Tdrkh
Gm42463
|
tudor and KH domain containing protein predicted gene 42463 |
| chr5_-_123038329 | 0.89 |
ENSMUST00000031435.14
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr11_+_78079562 | 0.89 |
ENSMUST00000108322.9
|
Rab34
|
RAB34, member RAS oncogene family |
| chr2_-_13496624 | 0.89 |
ENSMUST00000091436.7
|
Cubn
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr12_+_98234884 | 0.88 |
ENSMUST00000075072.6
|
Gpr65
|
G-protein coupled receptor 65 |
| chr8_+_107620251 | 0.88 |
ENSMUST00000212272.2
ENSMUST00000047629.7 |
Utp4
|
UTP4 small subunit processome component |
| chrX_+_84617624 | 0.87 |
ENSMUST00000048250.10
ENSMUST00000137438.2 ENSMUST00000146063.2 |
Tab3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr4_-_4138432 | 0.86 |
ENSMUST00000070375.8
|
Penk
|
preproenkephalin |
| chr7_-_43182595 | 0.86 |
ENSMUST00000205503.2
|
Cd33
|
CD33 antigen |
| chr19_-_8763609 | 0.86 |
ENSMUST00000177216.8
ENSMUST00000176610.9 ENSMUST00000177056.8 |
Taf6l
|
TATA-box binding protein associated factor 6 like |
| chr8_+_104828253 | 0.85 |
ENSMUST00000034339.10
|
Cdh5
|
cadherin 5 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 20.1 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 2.7 | 10.9 | GO:2001148 | positive regulation of cellular pH reduction(GO:0032849) dipeptide transmembrane transport(GO:0035442) regulation of oligopeptide transport(GO:0090088) regulation of dipeptide transport(GO:0090089) positive regulation of oligopeptide transport(GO:2000878) positive regulation of dipeptide transport(GO:2000880) regulation of dipeptide transmembrane transport(GO:2001148) positive regulation of dipeptide transmembrane transport(GO:2001150) |
| 2.6 | 7.9 | GO:0002780 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) neutrophil mediated killing of fungus(GO:0070947) |
| 2.0 | 6.1 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 1.7 | 5.2 | GO:0034118 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 1.4 | 4.3 | GO:0019542 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 1.4 | 4.3 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 1.3 | 3.9 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) |
| 1.3 | 5.1 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 1.1 | 3.3 | GO:1903632 | positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) |
| 1.1 | 4.3 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 1.0 | 3.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 1.0 | 3.0 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.9 | 2.8 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.9 | 5.5 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.9 | 4.5 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.9 | 2.7 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.9 | 4.3 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.8 | 4.6 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.8 | 2.3 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.7 | 2.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.7 | 4.8 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.7 | 2.7 | GO:0045575 | basophil activation(GO:0045575) |
| 0.6 | 14.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.6 | 7.0 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.6 | 3.4 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.5 | 1.6 | GO:0070376 | regulation of ERK5 cascade(GO:0070376) |
| 0.5 | 6.0 | GO:0008228 | opsonization(GO:0008228) |
| 0.5 | 1.6 | GO:0090172 | attachment of spindle microtubules to kinetochore involved in homologous chromosome segregation(GO:0051455) microtubule cytoskeleton organization involved in homologous chromosome segregation(GO:0090172) |
| 0.5 | 5.8 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.5 | 1.0 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.5 | 1.6 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.5 | 1.0 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.5 | 1.4 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 0.5 | 1.4 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.5 | 1.8 | GO:0033373 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 0.4 | 1.3 | GO:0042128 | nitrate assimilation(GO:0042128) |
| 0.4 | 2.6 | GO:0071899 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.4 | 9.1 | GO:0061615 | glycolytic process through fructose-6-phosphate(GO:0061615) |
| 0.4 | 1.2 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.4 | 1.2 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.4 | 3.7 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.4 | 1.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.3 | 1.0 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.3 | 3.5 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.3 | 3.0 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.3 | 3.0 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.3 | 0.6 | GO:1990051 | activation of protein kinase C activity(GO:1990051) |
| 0.3 | 1.2 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.3 | 2.8 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.3 | 2.7 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.3 | 1.8 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.3 | 1.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.3 | 0.8 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.3 | 0.8 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.3 | 4.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.3 | 6.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.3 | 0.8 | GO:0072248 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.3 | 1.0 | GO:2000409 | positive regulation of T cell extravasation(GO:2000409) |
| 0.2 | 0.7 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.2 | 1.7 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.2 | 1.0 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.2 | 1.9 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.2 | 1.6 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.2 | 1.4 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 0.9 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.2 | 4.0 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.2 | 0.7 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.2 | 1.3 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.2 | 4.0 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.2 | 4.4 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.2 | 0.8 | GO:0015786 | UDP-glucose transport(GO:0015786) |
| 0.2 | 1.0 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.2 | 1.9 | GO:0002870 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.2 | 1.7 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.2 | 1.5 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.2 | 2.9 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.2 | 1.4 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.2 | 3.3 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.2 | 3.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.2 | 1.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 1.7 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.2 | 0.8 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.2 | 0.8 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.2 | 0.5 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.2 | 1.0 | GO:0032831 | positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.2 | 1.6 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.2 | 1.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.2 | 0.5 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.2 | 0.9 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.2 | 1.1 | GO:0021747 | cochlear nucleus development(GO:0021747) |
| 0.1 | 1.0 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.1 | 0.9 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 1.5 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 1.9 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.1 | 3.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 2.7 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.1 | 0.5 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.1 | 0.4 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.1 | 1.3 | GO:0019661 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.1 | 0.6 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.1 | 0.4 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.1 | 0.4 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 0.4 | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.1 | 0.8 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 1.4 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.1 | 1.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.8 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.1 | 0.4 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 0.6 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.1 | 0.8 | GO:0051532 | regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.1 | 0.5 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 1.3 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 1.1 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 0.9 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.4 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.1 | 1.0 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.6 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 0.6 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 1.0 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.1 | 0.2 | GO:0002649 | regulation of tolerance induction to self antigen(GO:0002649) |
| 0.1 | 0.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.3 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.1 | 0.6 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.5 | GO:2001140 | regulation of phospholipid transport(GO:2001138) positive regulation of phospholipid transport(GO:2001140) |
| 0.1 | 0.6 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.7 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.9 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 1.3 | GO:0019377 | glycolipid catabolic process(GO:0019377) |
| 0.1 | 2.6 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.1 | 0.5 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.1 | 14.2 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 0.7 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.1 | 0.9 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.1 | 0.3 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.1 | 0.4 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.2 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.1 | 0.4 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 1.5 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.4 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 4.8 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 0.2 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.1 | 0.5 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.1 | 1.0 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 2.2 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.1 | 0.3 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.6 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 1.1 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 1.2 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 0.3 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) protein localization to site of double-strand break(GO:1990166) |
| 0.0 | 1.6 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.2 | GO:1990743 | protein sialylation(GO:1990743) negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 | 0.5 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.5 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 1.5 | GO:1903393 | positive regulation of focal adhesion assembly(GO:0051894) positive regulation of adherens junction organization(GO:1903393) |
| 0.0 | 0.4 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 1.0 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 1.5 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.2 | GO:0002266 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.1 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.0 | 0.3 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.6 | GO:1904867 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.6 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.3 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.0 | 0.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.6 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 1.3 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.3 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.9 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.3 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.1 | GO:0071042 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.5 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 1.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.8 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.8 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.5 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.7 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 1.0 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.4 | GO:0090500 | lymphatic endothelial cell differentiation(GO:0060836) endocardial cushion to mesenchymal transition(GO:0090500) |
| 0.0 | 0.3 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 1.4 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 1.6 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.6 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 1.3 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.1 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.0 | 0.1 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.5 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 2.7 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 1.2 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.1 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.8 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.2 | GO:0033085 | negative regulation of T cell differentiation in thymus(GO:0033085) |
| 0.0 | 0.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0045297 | mating plug formation(GO:0042628) single-organism reproductive behavior(GO:0044704) post-mating behavior(GO:0045297) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.2 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.4 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 0.3 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.0 | 1.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.6 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.5 | GO:0070571 | negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 1.1 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.5 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.2 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 1.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.1 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.2 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 2.2 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.5 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.2 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.2 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.3 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 | 0.2 | GO:0016045 | detection of bacterium(GO:0016045) detection of other organism(GO:0098543) |
| 0.0 | 1.0 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 2.5 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.5 | GO:0035196 | dsRNA fragmentation(GO:0031050) production of miRNAs involved in gene silencing by miRNA(GO:0035196) production of small RNA involved in gene silencing by RNA(GO:0070918) |
| 0.0 | 0.4 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.9 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 0.0 | 0.5 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 1.1 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.2 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.0 | 0.4 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.0 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) mesangial cell differentiation(GO:0072007) glomerular mesangial cell differentiation(GO:0072008) kidney interstitial fibroblast differentiation(GO:0072071) renal interstitial fibroblast development(GO:0072141) mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.0 | 0.1 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 1.2 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.7 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 14.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.7 | 4.4 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.5 | 5.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.5 | 2.8 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.5 | 1.4 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.3 | 6.1 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.3 | 4.0 | GO:0000796 | condensin complex(GO:0000796) |
| 0.3 | 2.9 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.3 | 7.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.3 | 0.9 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.3 | 0.9 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.3 | 4.9 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.3 | 2.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.3 | 1.3 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.3 | 1.8 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 6.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 1.2 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.2 | 0.9 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.2 | 3.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.2 | 0.8 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.2 | 0.9 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.2 | 1.0 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 1.8 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.2 | 4.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.2 | 12.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.9 | GO:0071547 | piP-body(GO:0071547) |
| 0.1 | 1.3 | GO:0045252 | dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 1.9 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 2.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 0.8 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 4.1 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 1.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 3.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.5 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 1.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 1.7 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 5.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 2.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.4 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 3.0 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 0.9 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 1.4 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 2.5 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 1.0 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.2 | GO:1904511 | cortical microtubule(GO:0055028) cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.1 | 2.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.1 | 0.6 | GO:0097422 | extrinsic component of endosome membrane(GO:0031313) tubular endosome(GO:0097422) |
| 0.1 | 1.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 9.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.7 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 2.3 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 0.4 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 1.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 1.0 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.5 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.1 | 7.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 0.5 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 0.6 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 1.7 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 3.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.7 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 2.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.2 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.6 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 1.1 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 3.0 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 1.4 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 5.9 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 1.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 3.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.0 | 0.9 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 3.1 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 1.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 12.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.8 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 9.0 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.7 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.9 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 1.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 2.2 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 2.2 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 1.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.8 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.0 | GO:1904602 | serotonin-activated cation-selective channel complex(GO:1904602) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 1.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 6.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 1.1 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 8.0 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 1.1 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 9.3 | GO:0005730 | nucleolus(GO:0005730) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 24.9 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 2.4 | 14.2 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 1.1 | 5.5 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 1.1 | 10.9 | GO:0004064 | arylesterase activity(GO:0004064) |
| 1.1 | 4.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 1.0 | 3.1 | GO:0004512 | inositol-3-phosphate synthase activity(GO:0004512) intramolecular lyase activity(GO:0016872) |
| 0.9 | 4.3 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.7 | 2.9 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.6 | 6.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.5 | 6.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.5 | 2.1 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.5 | 4.2 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.4 | 3.0 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.4 | 1.7 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.4 | 3.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.4 | 3.6 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.4 | 1.9 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.3 | 2.6 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.3 | 1.6 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.3 | 2.6 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.3 | 1.6 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.3 | 3.8 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.3 | 1.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.3 | 4.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.3 | 1.3 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.2 | 1.0 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.2 | 1.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 0.9 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.2 | 5.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 3.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.2 | 4.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.2 | 8.5 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.2 | 1.7 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 2.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 1.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 1.0 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.2 | 1.5 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.2 | 1.1 | GO:0030294 | receptor signaling protein tyrosine kinase inhibitor activity(GO:0030294) |
| 0.2 | 1.3 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.2 | 0.5 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.2 | 1.4 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.2 | 10.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.2 | 0.7 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.2 | 7.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 0.5 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 0.2 | 3.0 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 2.8 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 0.7 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 1.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 4.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.6 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 2.9 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 0.7 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.1 | 1.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 1.3 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.5 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 1.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.9 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 0.4 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 1.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.7 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 1.6 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 2.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.8 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 3.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.7 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.1 | 1.0 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 1.4 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 0.3 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.1 | 0.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.6 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 1.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 4.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 2.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.2 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.1 | 0.5 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.8 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.4 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 1.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.4 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 2.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.4 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 2.2 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 2.6 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.6 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.8 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.1 | 0.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 3.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.7 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 1.0 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.7 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 3.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.4 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.2 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 2.0 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 0.3 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 0.6 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.7 | GO:0001054 | RNA polymerase I activity(GO:0001054) RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.9 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.9 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 1.0 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 1.1 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.6 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.0 | 0.2 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.5 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.7 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 4.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 2.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 2.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 8.2 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.9 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.7 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 1.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.6 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 1.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 1.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 1.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 2.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 1.0 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.0 | 0.3 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 2.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.5 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 2.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 0.3 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.9 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.0 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.2 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 6.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.7 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 18.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 3.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 1.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.2 | 11.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 7.6 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 7.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 4.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.9 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 12.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 9.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 2.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 6.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 4.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 1.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 3.8 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 1.9 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 4.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 1.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 3.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 0.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 0.9 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 1.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 3.4 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 2.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 1.0 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 3.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.0 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 2.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.7 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.5 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.6 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.8 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 14.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.4 | 4.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 4.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 7.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 3.8 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.2 | 1.8 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 5.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.2 | 3.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 2.9 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 7.7 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.2 | 4.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 3.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 16.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 1.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 6.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.9 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 2.6 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.3 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 3.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 2.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 3.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 3.9 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 5.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 2.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 0.7 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 3.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 0.7 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 1.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 6.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 2.2 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 1.1 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.3 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 1.3 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.1 | 1.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 1.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 2.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 2.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 4.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.8 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.9 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 8.4 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 2.5 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 1.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.6 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.0 | 1.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.5 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 1.8 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 2.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.0 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.3 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.2 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.0 | 0.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.4 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |