Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Trp53
Z-value: 2.05
Transcription factors associated with Trp53
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Trp53
|
ENSMUSG00000059552.14 | Trp53 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Trp53 | mm39_v1_chr11_+_69471185_69471208 | 0.93 | 1.8e-16 | Click! |
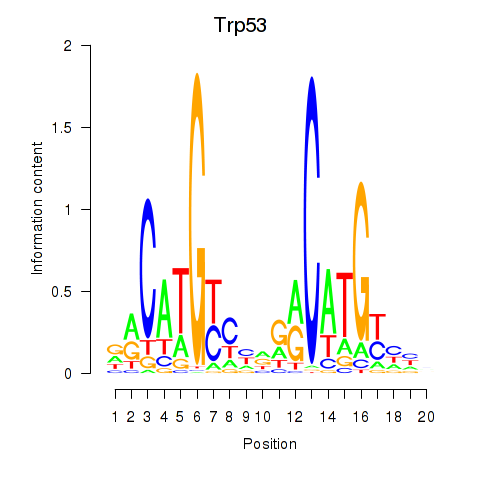
Activity profile of Trp53 motif
Sorted Z-values of Trp53 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Trp53
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_90576285 | 22.08 |
ENSMUST00000069927.10
|
S100a8
|
S100 calcium binding protein A8 (calgranulin A) |
| chr7_-_24459736 | 14.58 |
ENSMUST00000063956.7
|
Cd177
|
CD177 antigen |
| chr17_-_34109513 | 14.12 |
ENSMUST00000173386.2
ENSMUST00000114361.9 ENSMUST00000173492.9 |
Kifc1
|
kinesin family member C1 |
| chr8_+_95703728 | 12.21 |
ENSMUST00000179619.9
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr17_-_35285146 | 10.23 |
ENSMUST00000174190.2
ENSMUST00000097337.8 |
Mpig6b
|
megakaryocyte and platelet inhibitory receptor G6b |
| chr4_+_114914880 | 10.06 |
ENSMUST00000161601.8
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr17_+_28426752 | 8.53 |
ENSMUST00000002327.6
ENSMUST00000233560.2 ENSMUST00000233958.2 ENSMUST00000233170.2 |
Def6
|
differentially expressed in FDCP 6 |
| chr9_-_21874802 | 8.00 |
ENSMUST00000006397.7
|
Epor
|
erythropoietin receptor |
| chr17_+_36132567 | 7.98 |
ENSMUST00000003635.7
|
Ier3
|
immediate early response 3 |
| chr11_-_102815910 | 7.64 |
ENSMUST00000021311.10
|
Kif18b
|
kinesin family member 18B |
| chr6_-_136918885 | 7.31 |
ENSMUST00000111891.4
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr17_+_27136065 | 7.18 |
ENSMUST00000078961.6
|
Kifc5b
|
kinesin family member C5B |
| chr5_-_30278552 | 6.99 |
ENSMUST00000198095.2
ENSMUST00000196872.2 ENSMUST00000026846.11 |
Tyms
|
thymidylate synthase |
| chr2_-_69036489 | 6.99 |
ENSMUST00000127243.8
ENSMUST00000149643.2 ENSMUST00000167875.9 ENSMUST00000005365.15 |
Spc25
|
SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr6_-_136918495 | 6.99 |
ENSMUST00000111892.8
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr11_-_102360664 | 6.88 |
ENSMUST00000103086.4
|
Itga2b
|
integrin alpha 2b |
| chr6_-_136918844 | 6.45 |
ENSMUST00000204934.2
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr6_-_136918671 | 6.43 |
ENSMUST00000032344.12
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr8_+_95703506 | 6.30 |
ENSMUST00000212581.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr12_-_78953703 | 6.13 |
ENSMUST00000021544.8
|
Plek2
|
pleckstrin 2 |
| chr4_+_114914607 | 5.98 |
ENSMUST00000136946.8
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr10_+_75399920 | 5.88 |
ENSMUST00000141062.8
ENSMUST00000152657.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr8_-_22675773 | 5.59 |
ENSMUST00000046916.9
|
Ckap2
|
cytoskeleton associated protein 2 |
| chr2_-_69036472 | 5.52 |
ENSMUST00000112320.8
|
Spc25
|
SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr8_-_106578613 | 5.30 |
ENSMUST00000040776.6
|
Cenpt
|
centromere protein T |
| chr6_-_128868068 | 4.98 |
ENSMUST00000178918.2
ENSMUST00000160290.8 |
BC035044
|
cDNA sequence BC035044 |
| chr8_-_89362745 | 4.93 |
ENSMUST00000034087.9
|
Snx20
|
sorting nexin 20 |
| chr6_-_116437985 | 4.76 |
ENSMUST00000164547.8
ENSMUST00000170186.2 |
Alox5
|
arachidonate 5-lipoxygenase |
| chr15_+_85744147 | 4.66 |
ENSMUST00000231074.2
|
Gtse1
|
G two S phase expressed protein 1 |
| chr6_-_116438116 | 4.53 |
ENSMUST00000026795.13
|
Alox5
|
arachidonate 5-lipoxygenase |
| chr1_-_88133472 | 4.53 |
ENSMUST00000119972.4
|
Dnajb3
|
DnaJ heat shock protein family (Hsp40) member B3 |
| chr4_-_133981387 | 4.46 |
ENSMUST00000060050.6
|
Grrp1
|
glycine/arginine rich protein 1 |
| chr15_+_85743887 | 4.44 |
ENSMUST00000170629.3
|
Gtse1
|
G two S phase expressed protein 1 |
| chr3_-_132655804 | 4.40 |
ENSMUST00000117164.8
ENSMUST00000093971.5 ENSMUST00000042729.16 |
Npnt
|
nephronectin |
| chr17_+_28426831 | 4.33 |
ENSMUST00000233264.2
|
Def6
|
differentially expressed in FDCP 6 |
| chr5_+_35156389 | 4.16 |
ENSMUST00000114281.8
ENSMUST00000114280.8 |
Rgs12
|
regulator of G-protein signaling 12 |
| chr12_-_91351177 | 4.15 |
ENSMUST00000141429.8
|
Cep128
|
centrosomal protein 128 |
| chr8_+_70735477 | 4.04 |
ENSMUST00000087467.12
ENSMUST00000140212.8 ENSMUST00000110124.9 |
Homer3
|
homer scaffolding protein 3 |
| chr15_+_79784365 | 4.01 |
ENSMUST00000230135.2
|
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr15_-_102259158 | 4.00 |
ENSMUST00000231061.2
ENSMUST00000041208.9 |
Aaas
|
achalasia, adrenocortical insufficiency, alacrimia |
| chrX_+_73298388 | 3.84 |
ENSMUST00000119197.8
ENSMUST00000088313.5 |
Emd
|
emerin |
| chr11_+_58839716 | 3.81 |
ENSMUST00000078267.5
|
H2bu2
|
H2B.U histone 2 |
| chrX_+_73298342 | 3.81 |
ENSMUST00000096424.11
|
Emd
|
emerin |
| chr7_+_24584197 | 3.80 |
ENSMUST00000156372.8
ENSMUST00000124035.2 |
Rps19
|
ribosomal protein S19 |
| chr1_+_172327569 | 3.68 |
ENSMUST00000111230.8
|
Tagln2
|
transgelin 2 |
| chr18_-_35781422 | 3.47 |
ENSMUST00000237462.2
|
Mzb1
|
marginal zone B and B1 cell-specific protein 1 |
| chr17_+_29309942 | 3.44 |
ENSMUST00000119901.9
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chrX_+_73298285 | 3.27 |
ENSMUST00000002029.13
|
Emd
|
emerin |
| chr3_-_90297187 | 3.26 |
ENSMUST00000029541.12
|
Slc27a3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr14_-_56181993 | 3.20 |
ENSMUST00000022834.7
ENSMUST00000226280.2 |
Cma1
|
chymase 1, mast cell |
| chr13_+_94954202 | 2.99 |
ENSMUST00000220825.2
|
Tbca
|
tubulin cofactor A |
| chr7_+_24583994 | 2.82 |
ENSMUST00000108428.8
|
Rps19
|
ribosomal protein S19 |
| chr7_+_24584076 | 2.79 |
ENSMUST00000153451.9
ENSMUST00000108429.8 |
Rps19
|
ribosomal protein S19 |
| chr9_-_114811807 | 2.79 |
ENSMUST00000053150.8
|
Rps27rt
|
ribosomal protein S27, retrogene |
| chr11_-_79037213 | 2.72 |
ENSMUST00000018478.11
ENSMUST00000108264.8 |
Ksr1
|
kinase suppressor of ras 1 |
| chr7_-_141023199 | 2.61 |
ENSMUST00000106005.9
|
Pidd1
|
p53 induced death domain protein 1 |
| chr6_+_71684846 | 2.46 |
ENSMUST00000212792.2
|
Reep1
|
receptor accessory protein 1 |
| chr5_+_35146727 | 2.45 |
ENSMUST00000114284.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr1_+_75377616 | 2.44 |
ENSMUST00000122266.3
|
Speg
|
SPEG complex locus |
| chr7_+_89780785 | 2.37 |
ENSMUST00000208684.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr11_-_115968373 | 2.35 |
ENSMUST00000174822.8
|
Unc13d
|
unc-13 homolog D |
| chr10_+_110581293 | 2.34 |
ENSMUST00000174857.8
ENSMUST00000073781.12 ENSMUST00000173471.8 ENSMUST00000173634.2 |
E2f7
|
E2F transcription factor 7 |
| chr5_+_35156454 | 2.31 |
ENSMUST00000114283.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr1_+_82817794 | 2.21 |
ENSMUST00000186043.2
|
Agfg1
|
ArfGAP with FG repeats 1 |
| chr5_+_144127102 | 2.19 |
ENSMUST00000060747.8
|
Bhlha15
|
basic helix-loop-helix family, member a15 |
| chr5_+_35146880 | 2.08 |
ENSMUST00000114285.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr6_-_38814159 | 2.04 |
ENSMUST00000160962.8
|
Hipk2
|
homeodomain interacting protein kinase 2 |
| chr13_-_98951890 | 2.02 |
ENSMUST00000040340.16
ENSMUST00000179563.8 ENSMUST00000109403.2 |
Fcho2
|
FCH domain only 2 |
| chr11_-_3477916 | 1.96 |
ENSMUST00000020718.10
|
Smtn
|
smoothelin |
| chr2_+_128942900 | 1.94 |
ENSMUST00000103205.11
|
Polr1b
|
polymerase (RNA) I polypeptide B |
| chr13_-_22227114 | 1.92 |
ENSMUST00000091741.6
|
H2ac11
|
H2A clustered histone 11 |
| chr9_-_110886576 | 1.89 |
ENSMUST00000199839.5
|
Ccrl2
|
chemokine (C-C motif) receptor-like 2 |
| chr6_-_29212295 | 1.82 |
ENSMUST00000078155.12
|
Impdh1
|
inosine monophosphate dehydrogenase 1 |
| chr14_-_73563212 | 1.82 |
ENSMUST00000022701.7
|
Rb1
|
RB transcriptional corepressor 1 |
| chr11_+_118367651 | 1.81 |
ENSMUST00000135383.9
|
Engase
|
endo-beta-N-acetylglucosaminidase |
| chrX_-_141089165 | 1.73 |
ENSMUST00000134825.3
|
Kcne1l
|
potassium voltage-gated channel, Isk-related family, member 1-like, pseudogene |
| chr11_-_116058961 | 1.71 |
ENSMUST00000139020.2
ENSMUST00000103031.8 ENSMUST00000124828.8 |
Fbf1
|
Fas (TNFRSF6) binding factor 1 |
| chr2_+_128942919 | 1.70 |
ENSMUST00000028874.8
|
Polr1b
|
polymerase (RNA) I polypeptide B |
| chr13_+_22227359 | 1.68 |
ENSMUST00000110452.2
|
H2bc11
|
H2B clustered histone 11 |
| chr1_+_133278248 | 1.68 |
ENSMUST00000094556.3
|
Ren1
|
renin 1 structural |
| chr6_+_41098273 | 1.63 |
ENSMUST00000103270.4
|
Trbv13-2
|
T cell receptor beta, variable 13-2 |
| chr9_+_5298669 | 1.62 |
ENSMUST00000238505.2
|
Casp1
|
caspase 1 |
| chr1_+_51328265 | 1.59 |
ENSMUST00000051572.8
|
Cavin2
|
caveolae associated 2 |
| chr4_-_135300934 | 1.58 |
ENSMUST00000105855.2
|
Grhl3
|
grainyhead like transcription factor 3 |
| chr6_+_71684872 | 1.51 |
ENSMUST00000212631.2
|
Reep1
|
receptor accessory protein 1 |
| chr9_-_56151334 | 1.48 |
ENSMUST00000188142.7
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr4_-_129534853 | 1.47 |
ENSMUST00000046425.16
ENSMUST00000133803.8 |
Txlna
|
taxilin alpha |
| chr1_-_38704028 | 1.43 |
ENSMUST00000039827.14
|
Aff3
|
AF4/FMR2 family, member 3 |
| chr13_-_23755374 | 1.42 |
ENSMUST00000102969.6
|
H2ac8
|
H2A clustered histone 8 |
| chr6_+_127423779 | 1.38 |
ENSMUST00000112191.8
|
Parp11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr3_-_19319155 | 1.38 |
ENSMUST00000091314.11
|
Pde7a
|
phosphodiesterase 7A |
| chr4_-_129534403 | 1.37 |
ENSMUST00000084264.12
|
Txlna
|
taxilin alpha |
| chr6_-_124698805 | 1.35 |
ENSMUST00000173315.8
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr4_-_129534752 | 1.34 |
ENSMUST00000132217.8
ENSMUST00000130017.2 ENSMUST00000154105.8 |
Txlna
|
taxilin alpha |
| chr16_+_20536415 | 1.28 |
ENSMUST00000021405.8
|
Polr2h
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr14_-_8536869 | 1.26 |
ENSMUST00000238865.2
ENSMUST00000159275.4 |
Gm11100
|
predicted gene 11100 |
| chr7_+_75105282 | 1.26 |
ENSMUST00000207750.2
ENSMUST00000166315.7 |
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr8_-_112580910 | 1.25 |
ENSMUST00000034432.7
|
Cfdp1
|
craniofacial development protein 1 |
| chr2_+_29780073 | 1.21 |
ENSMUST00000028128.13
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr16_+_20536545 | 1.17 |
ENSMUST00000231656.2
|
Polr2h
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr5_+_114845821 | 1.17 |
ENSMUST00000094441.11
|
Tchp
|
trichoplein, keratin filament binding |
| chr6_+_67586695 | 1.14 |
ENSMUST00000103303.3
|
Igkv1-135
|
immunoglobulin kappa variable 1-135 |
| chr2_-_163239865 | 1.13 |
ENSMUST00000017961.11
ENSMUST00000109425.3 |
Jph2
|
junctophilin 2 |
| chr13_-_98152768 | 1.08 |
ENSMUST00000238746.2
|
Arhgef28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr1_-_192812509 | 1.06 |
ENSMUST00000085555.7
|
Utp25
|
UTP25 small subunit processome component |
| chr1_-_79836344 | 1.02 |
ENSMUST00000027467.11
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr2_+_85838122 | 1.01 |
ENSMUST00000062166.2
|
Olfr1032
|
olfactory receptor 1032 |
| chr13_+_22220000 | 1.00 |
ENSMUST00000110455.4
|
H2bc12
|
H2B clustered histone 12 |
| chr13_-_74956640 | 0.99 |
ENSMUST00000231578.2
|
Cast
|
calpastatin |
| chr9_-_106324642 | 0.98 |
ENSMUST00000185334.7
ENSMUST00000187001.2 ENSMUST00000171678.9 ENSMUST00000190798.7 ENSMUST00000048685.13 ENSMUST00000171925.8 |
Abhd14a
|
abhydrolase domain containing 14A |
| chr11_+_6366259 | 0.96 |
ENSMUST00000213200.2
|
Ppia
|
peptidylprolyl isomerase A |
| chr14_-_70412804 | 0.95 |
ENSMUST00000143393.2
|
Pdlim2
|
PDZ and LIM domain 2 |
| chr13_-_22219738 | 0.95 |
ENSMUST00000091742.6
|
H2ac12
|
H2A clustered histone 12 |
| chr8_+_18896267 | 0.93 |
ENSMUST00000149565.8
ENSMUST00000033847.5 |
Agpat5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon) |
| chr4_-_111755892 | 0.92 |
ENSMUST00000102720.8
|
Slc5a9
|
solute carrier family 5 (sodium/glucose cotransporter), member 9 |
| chr14_-_49303826 | 0.92 |
ENSMUST00000161504.8
|
Exoc5
|
exocyst complex component 5 |
| chr15_-_102154874 | 0.89 |
ENSMUST00000063339.14
|
Rarg
|
retinoic acid receptor, gamma |
| chr17_-_22152003 | 0.87 |
ENSMUST00000074295.8
|
Zfp942
|
zinc finger protein 942 |
| chr5_-_6926523 | 0.85 |
ENSMUST00000164784.2
|
Zfp804b
|
zinc finger protein 804B |
| chr19_+_46316617 | 0.85 |
ENSMUST00000026256.9
ENSMUST00000177667.2 |
Fbxl15
|
F-box and leucine-rich repeat protein 15 |
| chr19_-_47126719 | 0.83 |
ENSMUST00000140512.8
ENSMUST00000035822.2 |
Calhm2
|
calcium homeostasis modulator family member 2 |
| chr11_+_55360502 | 0.76 |
ENSMUST00000018727.4
|
G3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr7_+_24099037 | 0.74 |
ENSMUST00000002280.11
|
Smg9
|
smg-9 homolog, nonsense mediated mRNA decay factor (C. elegans) |
| chr3_+_116653113 | 0.74 |
ENSMUST00000040260.11
|
Frrs1
|
ferric-chelate reductase 1 |
| chr10_+_94386714 | 0.73 |
ENSMUST00000148910.3
ENSMUST00000117460.8 |
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr1_-_181985663 | 0.73 |
ENSMUST00000169123.4
|
Vmn1r1
|
vomeronasal 1 receptor 1 |
| chr13_-_55718899 | 0.70 |
ENSMUST00000225240.3
ENSMUST00000021957.8 |
Fam193b
|
family with sequence similarity 193, member B |
| chr4_-_9643636 | 0.70 |
ENSMUST00000108333.8
ENSMUST00000108334.8 ENSMUST00000108335.8 ENSMUST00000152526.8 ENSMUST00000103004.10 |
Asph
|
aspartate-beta-hydroxylase |
| chr1_+_171386752 | 0.70 |
ENSMUST00000004829.13
|
Cd244a
|
CD244 molecule A |
| chr2_-_91795910 | 0.67 |
ENSMUST00000239257.2
|
Dgkz
|
diacylglycerol kinase zeta |
| chr7_-_64522741 | 0.66 |
ENSMUST00000094331.5
|
Nsmce3
|
NSE3 homolog, SMC5-SMC6 complex component |
| chr18_+_47245204 | 0.62 |
ENSMUST00000234633.2
|
Hspe1-rs1
|
heat shock protein 1 (chaperonin 10), related sequence 1 |
| chr9_-_106035308 | 0.62 |
ENSMUST00000159809.2
ENSMUST00000162562.2 ENSMUST00000036382.13 |
Glyctk
|
glycerate kinase |
| chr2_-_155434487 | 0.61 |
ENSMUST00000155347.2
ENSMUST00000130881.8 ENSMUST00000079691.13 |
Gss
|
glutathione synthetase |
| chr9_-_19968858 | 0.59 |
ENSMUST00000216538.2
ENSMUST00000212098.3 |
Olfr867
|
olfactory receptor 867 |
| chr15_-_77854988 | 0.58 |
ENSMUST00000100484.6
|
Eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr16_+_38279289 | 0.58 |
ENSMUST00000099816.3
ENSMUST00000232409.2 |
Cd80
|
CD80 antigen |
| chr15_-_77854711 | 0.58 |
ENSMUST00000230419.2
|
Eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr3_+_59832635 | 0.58 |
ENSMUST00000049476.3
|
Aadacl2fm1
|
AADACL2 family member 1 |
| chr9_-_106035332 | 0.54 |
ENSMUST00000112543.9
|
Glyctk
|
glycerate kinase |
| chr13_+_21938258 | 0.54 |
ENSMUST00000091709.3
|
H2bc15
|
H2B clustered histone 15 |
| chr7_-_28661648 | 0.53 |
ENSMUST00000127210.8
|
Actn4
|
actinin alpha 4 |
| chr13_+_23758555 | 0.52 |
ENSMUST00000090776.7
|
H2ac7
|
H2A clustered histone 7 |
| chr12_-_114330574 | 0.50 |
ENSMUST00000103485.3
|
Ighv12-3
|
immunoglobulin heavy variable V12-3 |
| chr10_+_56253418 | 0.49 |
ENSMUST00000068581.9
ENSMUST00000217789.2 |
Gja1
|
gap junction protein, alpha 1 |
| chr1_+_161796854 | 0.48 |
ENSMUST00000160881.2
ENSMUST00000159648.2 |
Pigc
|
phosphatidylinositol glycan anchor biosynthesis, class C |
| chr7_+_103586589 | 0.47 |
ENSMUST00000098189.4
|
Olfr632
|
olfactory receptor 632 |
| chr15_+_100052260 | 0.46 |
ENSMUST00000023768.14
|
Dip2b
|
disco interacting protein 2 homolog B |
| chr9_-_58065800 | 0.46 |
ENSMUST00000168864.4
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr1_+_161796748 | 0.45 |
ENSMUST00000111594.3
ENSMUST00000028021.7 ENSMUST00000193784.2 ENSMUST00000161826.2 |
Pigc
Gm38304
|
phosphatidylinositol glycan anchor biosynthesis, class C predicted gene, 38304 |
| chr10_+_78410803 | 0.45 |
ENSMUST00000218763.2
ENSMUST00000220430.2 ENSMUST00000218885.2 ENSMUST00000218215.2 ENSMUST00000218271.2 |
Ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr3_-_92528480 | 0.43 |
ENSMUST00000170676.3
|
Lce6a
|
late cornified envelope 6A |
| chr16_+_20470402 | 0.43 |
ENSMUST00000007212.9
ENSMUST00000232629.2 |
Psmd2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr10_-_129962924 | 0.39 |
ENSMUST00000074161.2
|
Olfr824
|
olfactory receptor 824 |
| chr11_-_32217547 | 0.38 |
ENSMUST00000109389.9
ENSMUST00000129010.2 ENSMUST00000020530.12 |
Nprl3
|
nitrogen permease regulator-like 3 |
| chr11_+_81948649 | 0.35 |
ENSMUST00000000342.3
|
Ccl11
|
chemokine (C-C motif) ligand 11 |
| chr5_+_136086045 | 0.34 |
ENSMUST00000111137.8
ENSMUST00000156530.8 ENSMUST00000006303.11 |
Upk3bl
|
uroplakin 3B-like |
| chr1_-_160958998 | 0.33 |
ENSMUST00000111611.8
|
Klhl20
|
kelch-like 20 |
| chr2_+_29779227 | 0.32 |
ENSMUST00000123335.8
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr1_+_40364752 | 0.32 |
ENSMUST00000193388.2
|
Il1rl2
|
interleukin 1 receptor-like 2 |
| chr7_+_24099346 | 0.31 |
ENSMUST00000108434.2
|
Smg9
|
smg-9 homolog, nonsense mediated mRNA decay factor (C. elegans) |
| chr9_+_65253374 | 0.31 |
ENSMUST00000048184.4
ENSMUST00000214433.2 |
Pdcd7
|
programmed cell death 7 |
| chr14_-_9015639 | 0.30 |
ENSMUST00000112656.4
|
Synpr
|
synaptoporin |
| chr12_-_26465253 | 0.30 |
ENSMUST00000020971.14
|
Rnf144a
|
ring finger protein 144A |
| chr5_-_135494775 | 0.29 |
ENSMUST00000212301.2
|
Hip1
|
huntingtin interacting protein 1 |
| chr7_-_28661751 | 0.27 |
ENSMUST00000068045.14
ENSMUST00000217157.2 |
Actn4
|
actinin alpha 4 |
| chr6_+_90246088 | 0.27 |
ENSMUST00000058039.3
|
Vmn1r54
|
vomeronasal 1 receptor 54 |
| chr9_+_45049687 | 0.26 |
ENSMUST00000060125.7
|
Scn4b
|
sodium channel, type IV, beta |
| chr6_+_68414401 | 0.26 |
ENSMUST00000103324.3
|
Igkv15-103
|
immunoglobulin kappa chain variable 15-103 |
| chr7_-_3828640 | 0.26 |
ENSMUST00000189095.7
ENSMUST00000094911.5 ENSMUST00000153846.8 ENSMUST00000108619.8 ENSMUST00000108620.8 |
Gm15448
|
predicted gene 15448 |
| chr9_-_121621544 | 0.25 |
ENSMUST00000035110.11
|
Hhatl
|
hedgehog acyltransferase-like |
| chr5_+_138019523 | 0.24 |
ENSMUST00000171498.3
ENSMUST00000085886.3 |
Smok3a
|
sperm motility kinase 3A |
| chr16_+_16962464 | 0.23 |
ENSMUST00000231726.2
|
Ydjc
|
YdjC homolog (bacterial) |
| chr15_-_73114855 | 0.23 |
ENSMUST00000227686.2
|
Ptk2
|
PTK2 protein tyrosine kinase 2 |
| chr2_+_72115981 | 0.23 |
ENSMUST00000090824.12
ENSMUST00000135469.8 |
Map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr4_-_14796052 | 0.22 |
ENSMUST00000108276.2
ENSMUST00000023917.8 |
Lrrc69
|
leucine rich repeat containing 69 |
| chr1_+_55276336 | 0.21 |
ENSMUST00000061334.10
|
Mars2
|
methionine-tRNA synthetase 2 (mitochondrial) |
| chr2_+_37029334 | 0.21 |
ENSMUST00000214905.2
ENSMUST00000217298.2 ENSMUST00000104995.3 |
Olfr364-ps1
|
olfactory receptor 364, pseudogene 1 |
| chr19_-_45580258 | 0.19 |
ENSMUST00000160003.8
ENSMUST00000162879.8 |
Fbxw4
|
F-box and WD-40 domain protein 4 |
| chr9_+_38630317 | 0.19 |
ENSMUST00000129598.2
|
Vwa5a
|
von Willebrand factor A domain containing 5A |
| chr4_+_155875629 | 0.18 |
ENSMUST00000105593.2
|
Ankrd65
|
ankyrin repeat domain 65 |
| chr3_+_20043315 | 0.15 |
ENSMUST00000173779.2
|
Cp
|
ceruloplasmin |
| chr6_+_68547717 | 0.15 |
ENSMUST00000196839.5
ENSMUST00000103327.3 |
Igkv12-98
|
immunoglobulin kappa variable 12-98 |
| chr4_+_148642879 | 0.15 |
ENSMUST00000017408.14
ENSMUST00000076022.7 |
Exosc10
|
exosome component 10 |
| chr11_+_88095222 | 0.14 |
ENSMUST00000118784.8
ENSMUST00000139170.8 ENSMUST00000107915.10 ENSMUST00000144070.3 |
Mrps23
|
mitochondrial ribosomal protein S23 |
| chr19_-_5399368 | 0.14 |
ENSMUST00000238111.2
|
Cst6
|
cystatin E/M |
| chr5_+_36361360 | 0.14 |
ENSMUST00000052224.6
|
Psapl1
|
prosaposin-like 1 |
| chr14_+_73475335 | 0.13 |
ENSMUST00000044405.8
|
Lpar6
|
lysophosphatidic acid receptor 6 |
| chr10_+_21469039 | 0.12 |
ENSMUST00000057341.6
|
1700020N01Rik
|
RIKEN cDNA 1700020N01 gene |
| chr12_-_26465130 | 0.12 |
ENSMUST00000222082.2
ENSMUST00000062149.6 |
Rnf144a
|
ring finger protein 144A |
| chr5_-_138270995 | 0.12 |
ENSMUST00000161665.2
ENSMUST00000100530.8 ENSMUST00000161279.8 ENSMUST00000161647.8 |
Gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr7_-_18301211 | 0.12 |
ENSMUST00000094794.5
|
Psg27
|
pregnancy-specific glycoprotein 27 |
| chr19_-_10581622 | 0.11 |
ENSMUST00000037678.7
|
Tkfc
|
triokinase, FMN cyclase |
| chr11_-_59078565 | 0.10 |
ENSMUST00000170895.3
|
Guk1
|
guanylate kinase 1 |
| chr2_+_37080286 | 0.10 |
ENSMUST00000218602.2
|
Olfr365
|
olfactory receptor 365 |
| chr3_-_116474909 | 0.09 |
ENSMUST00000140672.3
|
Gm43191
|
predicted gene 43191 |
| chr9_+_121946321 | 0.09 |
ENSMUST00000119215.9
ENSMUST00000146832.8 ENSMUST00000118886.9 ENSMUST00000120173.9 ENSMUST00000139181.2 |
Snrk
|
SNF related kinase |
| chr14_-_104081119 | 0.08 |
ENSMUST00000227824.2
ENSMUST00000172237.2 |
Ednrb
|
endothelin receptor type B |
| chr4_+_102287244 | 0.07 |
ENSMUST00000172616.2
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr11_+_66802807 | 0.07 |
ENSMUST00000123434.3
|
Pirt
|
phosphoinositide-interacting regulator of transient receptor potential channels |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 22.1 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 5.3 | 16.0 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 3.5 | 14.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 3.0 | 27.2 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 2.4 | 9.4 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 2.2 | 11.0 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 1.5 | 9.3 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 1.4 | 7.0 | GO:0046073 | dTMP biosynthetic process(GO:0006231) dTMP metabolic process(GO:0046073) |
| 1.0 | 4.2 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.9 | 3.6 | GO:0017126 | nucleologenesis(GO:0017126) |
| 0.9 | 8.0 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.9 | 4.4 | GO:0097195 | pilomotor reflex(GO:0097195) positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.8 | 3.2 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 0.7 | 18.5 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.7 | 5.9 | GO:0031179 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.7 | 3.5 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.6 | 3.0 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.5 | 8.0 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.5 | 10.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.4 | 1.6 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.4 | 2.4 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.3 | 1.0 | GO:0044704 | mating plug formation(GO:0042628) single-organism reproductive behavior(GO:0044704) post-mating behavior(GO:0045297) seminal vesicle development(GO:0061107) |
| 0.3 | 6.1 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.3 | 5.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.3 | 2.3 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.3 | 1.2 | GO:0002191 | formation of cytoplasmic translation initiation complex(GO:0001732) cap-dependent translational initiation(GO:0002191) |
| 0.3 | 1.7 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.3 | 1.8 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.2 | 0.7 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.2 | 2.0 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.2 | 0.8 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.2 | 4.0 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.2 | 0.9 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.2 | 1.4 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.2 | 3.5 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.2 | 0.5 | GO:0003347 | atrial ventricular junction remodeling(GO:0003294) epicardial cell to mesenchymal cell transition(GO:0003347) regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.2 | 12.4 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.2 | 1.3 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.2 | 4.0 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 1.7 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.1 | 1.1 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 | 1.2 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 0.2 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.1 | 1.6 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 1.8 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 2.2 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.1 | 1.3 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 2.2 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 11.8 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.1 | 2.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 6.9 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 0.9 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 1.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 1.0 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 1.0 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 1.7 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 1.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 6.1 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.7 | GO:0072711 | cellular response to hydroxyurea(GO:0072711) |
| 0.0 | 0.2 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 5.3 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.6 | GO:0046641 | positive regulation of alpha-beta T cell proliferation(GO:0046641) |
| 0.0 | 0.7 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 2.1 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 1.0 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.1 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 1.4 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 0.3 | GO:0002551 | mast cell chemotaxis(GO:0002551) |
| 0.0 | 3.0 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 0.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0009153 | purine deoxyribonucleotide biosynthetic process(GO:0009153) |
| 0.0 | 0.1 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 2.3 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.9 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.0 | GO:2001201 | regulation of transforming growth factor-beta secretion(GO:2001201) |
| 0.0 | 1.0 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.3 | GO:0099623 | regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) regulation of cardiac muscle cell membrane repolarization(GO:0099623) |
| 0.0 | 0.6 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 1.5 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.9 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.4 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 16.0 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 2.1 | 12.5 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 1.7 | 18.5 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 1.1 | 3.4 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 1.1 | 11.6 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.7 | 14.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.7 | 7.6 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.5 | 9.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.5 | 2.0 | GO:0098833 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.5 | 4.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.4 | 5.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.4 | 6.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.4 | 1.6 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.4 | 1.8 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.3 | 2.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.2 | 2.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.2 | 3.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 11.0 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.2 | 3.5 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.2 | 14.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 4.0 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 6.9 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 8.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.4 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 1.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 1.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 7.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 17.3 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.1 | 0.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 1.0 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 0.7 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.3 | GO:0098890 | extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 12.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.9 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 3.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 5.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.2 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 4.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 3.0 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 2.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 2.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 7.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 1.6 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.3 | GO:0000776 | kinetochore(GO:0000776) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 27.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 2.8 | 22.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 1.9 | 9.3 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 1.4 | 6.9 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 1.1 | 3.4 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.6 | 3.5 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.6 | 4.0 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.5 | 7.0 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.5 | 5.9 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.5 | 6.1 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.4 | 12.9 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.4 | 1.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.4 | 3.3 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.3 | 0.3 | GO:0019961 | interferon binding(GO:0019961) |
| 0.3 | 1.8 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.2 | 18.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 9.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.2 | 4.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 16.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 1.8 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.2 | 12.7 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 7.6 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 4.0 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.2 | 14.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.2 | 2.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 2.0 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 9.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 4.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.0 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 1.0 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 1.6 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 1.7 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 0.3 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 1.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.7 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.3 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 0.9 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.9 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 3.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 8.3 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 1.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.9 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 1.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.9 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.7 | GO:0001727 | lipid kinase activity(GO:0001727) diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.9 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 4.2 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.3 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.4 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0034012 | glycerone kinase activity(GO:0004371) FAD-AMP lyase (cyclizing) activity(GO:0034012) triokinase activity(GO:0050354) |
| 0.0 | 0.9 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.6 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 3.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 2.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 5.3 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 3.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.7 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 21.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.4 | 40.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.3 | 3.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 6.9 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 9.4 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 11.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 4.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 1.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 2.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.1 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.9 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 2.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.2 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.9 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 14.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.3 | 7.1 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.3 | 7.0 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 1.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.2 | 6.1 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.2 | 1.8 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.2 | 17.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 10.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 3.0 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 4.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 6.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 3.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 27.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.8 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 3.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.4 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.6 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 9.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 2.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.1 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |