Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Trp63
Z-value: 0.58
Transcription factors associated with Trp63
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Trp63
|
ENSMUSG00000022510.15 | Trp63 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Trp63 | mm39_v1_chr16_+_25620652_25620678 | 0.01 | 9.6e-01 | Click! |
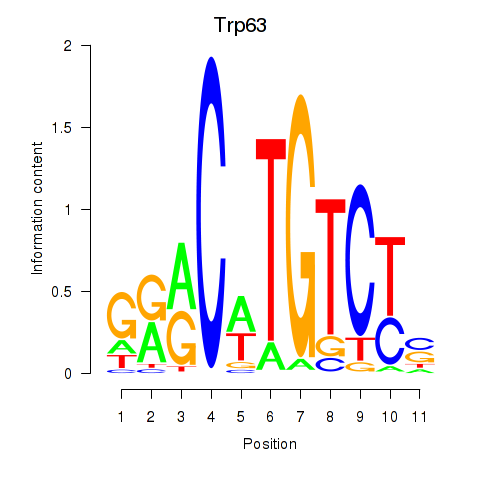
Activity profile of Trp63 motif
Sorted Z-values of Trp63 motif
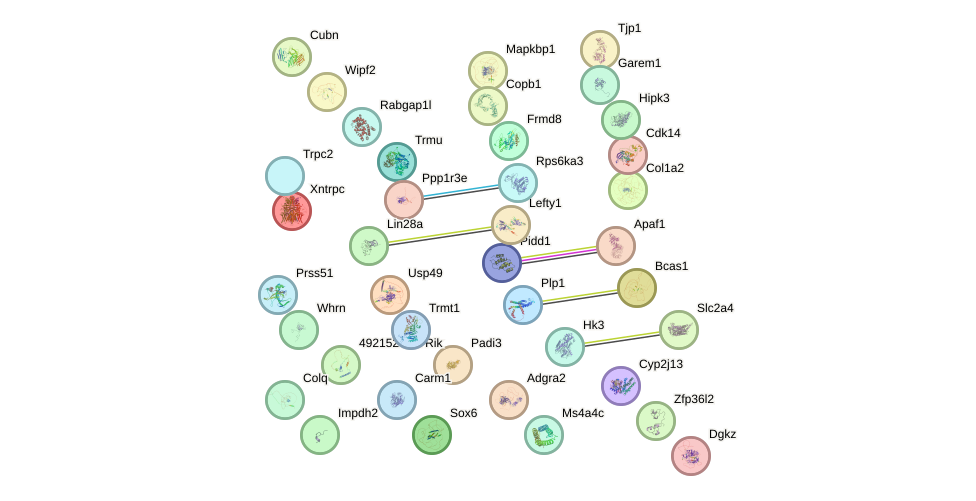
Network of associatons between targets according to the STRING database.
First level regulatory network of Trp63
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_69838971 | 1.79 |
ENSMUST00000179298.3
ENSMUST00000018710.13 ENSMUST00000135437.3 ENSMUST00000141837.9 ENSMUST00000142500.8 |
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr7_-_141023199 | 1.73 |
ENSMUST00000106005.9
|
Pidd1
|
p53 induced death domain protein 1 |
| chr7_-_115637970 | 1.68 |
ENSMUST00000166877.8
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr2_-_13496624 | 1.62 |
ENSMUST00000091436.7
|
Cubn
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr2_-_170269748 | 1.47 |
ENSMUST00000013667.3
ENSMUST00000109152.9 ENSMUST00000068137.11 |
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr17_+_47983587 | 1.35 |
ENSMUST00000152724.2
|
Usp49
|
ubiquitin specific peptidase 49 |
| chr19_+_53128901 | 1.24 |
ENSMUST00000235754.2
ENSMUST00000237301.2 ENSMUST00000238130.2 |
Add3
|
adducin 3 (gamma) |
| chr1_+_180762587 | 1.21 |
ENSMUST00000037361.9
|
Lefty1
|
left right determination factor 1 |
| chr9_+_108437485 | 1.11 |
ENSMUST00000081111.14
ENSMUST00000193421.2 |
Impdh2
|
inosine monophosphate dehydrogenase 2 |
| chr13_-_55169100 | 1.05 |
ENSMUST00000148221.8
ENSMUST00000052949.13 |
Hk3
|
hexokinase 3 |
| chr13_-_55169000 | 0.96 |
ENSMUST00000153665.8
|
Hk3
|
hexokinase 3 |
| chr10_-_90918566 | 0.95 |
ENSMUST00000162618.8
ENSMUST00000020157.13 ENSMUST00000160788.2 |
Apaf1
|
apoptotic peptidase activating factor 1 |
| chr19_+_53128861 | 0.73 |
ENSMUST00000111741.10
|
Add3
|
adducin 3 (gamma) |
| chr6_+_34686543 | 0.72 |
ENSMUST00000031775.13
|
Cald1
|
caldesmon 1 |
| chrX_+_158038778 | 0.66 |
ENSMUST00000126686.8
ENSMUST00000033671.13 |
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr18_-_21433186 | 0.58 |
ENSMUST00000049260.7
|
Garem1
|
GRB2 associated regulator of MAPK1 subtype 1 |
| chr1_-_160134873 | 0.56 |
ENSMUST00000193185.6
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr7_-_125760164 | 0.50 |
ENSMUST00000164741.2
|
Xpo6
|
exportin 6 |
| chr19_-_5925239 | 0.50 |
ENSMUST00000155227.2
|
Frmd8
|
FERM domain containing 8 |
| chr6_+_4505493 | 0.46 |
ENSMUST00000031668.10
|
Col1a2
|
collagen, type I, alpha 2 |
| chr4_-_133746138 | 0.46 |
ENSMUST00000051674.3
|
Lin28a
|
lin-28 homolog A (C. elegans) |
| chr8_+_85412928 | 0.43 |
ENSMUST00000177531.8
|
Trmt1
|
tRNA methyltransferase 1 |
| chr18_-_21433167 | 0.40 |
ENSMUST00000234115.2
|
Garem1
|
GRB2 associated regulator of MAPK1 subtype 1 |
| chr19_-_40365318 | 0.34 |
ENSMUST00000239304.2
|
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr19_+_11382092 | 0.34 |
ENSMUST00000153546.8
|
Ms4a4c
|
membrane-spanning 4-domains, subfamily A, member 4C |
| chr8_-_86159398 | 0.31 |
ENSMUST00000047749.7
|
4921524J17Rik
|
RIKEN cDNA 4921524J17 gene |
| chr19_-_5925296 | 0.31 |
ENSMUST00000025728.13
|
Frmd8
|
FERM domain containing 8 |
| chr8_+_27575611 | 0.31 |
ENSMUST00000178514.8
ENSMUST00000033876.14 |
Adgra2
|
adhesion G protein-coupled receptor A2 |
| chr2_-_91795910 | 0.29 |
ENSMUST00000239257.2
|
Dgkz
|
diacylglycerol kinase zeta |
| chr5_-_5315968 | 0.29 |
ENSMUST00000115451.8
ENSMUST00000115452.8 ENSMUST00000131392.8 |
Cdk14
|
cyclin-dependent kinase 14 |
| chr9_+_21498591 | 0.27 |
ENSMUST00000216160.2
|
Carm1
|
coactivator-associated arginine methyltransferase 1 |
| chr7_+_24584197 | 0.24 |
ENSMUST00000156372.8
ENSMUST00000124035.2 |
Rps19
|
ribosomal protein S19 |
| chr17_+_29309942 | 0.23 |
ENSMUST00000119901.9
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chr14_-_31299275 | 0.22 |
ENSMUST00000112027.9
|
Colq
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr17_-_84495364 | 0.19 |
ENSMUST00000060366.7
|
Zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr6_+_11907808 | 0.18 |
ENSMUST00000155037.4
|
Phf14
|
PHD finger protein 14 |
| chr12_-_114646685 | 0.17 |
ENSMUST00000194350.6
ENSMUST00000103504.3 |
Ighv1-18
|
immunoglobulin heavy variable V1-18 |
| chr7_-_113853894 | 0.17 |
ENSMUST00000033012.9
|
Copb1
|
coatomer protein complex, subunit beta 1 |
| chr4_-_140537957 | 0.14 |
ENSMUST00000026377.9
|
Padi3
|
peptidyl arginine deiminase, type III |
| chr14_+_64331130 | 0.14 |
ENSMUST00000224112.2
ENSMUST00000165710.2 ENSMUST00000170709.2 |
Prss51
|
protease, serine 51 |
| chr7_-_65020655 | 0.11 |
ENSMUST00000032729.8
|
Tjp1
|
tight junction protein 1 |
| chr2_+_119803230 | 0.11 |
ENSMUST00000229024.2
|
Mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr11_+_98754434 | 0.10 |
ENSMUST00000142414.8
ENSMUST00000037480.9 |
Wipf2
|
WAS/WASL interacting protein family, member 2 |
| chr7_+_24583994 | 0.09 |
ENSMUST00000108428.8
|
Rps19
|
ribosomal protein S19 |
| chr6_+_34686373 | 0.07 |
ENSMUST00000115021.8
|
Cald1
|
caldesmon 1 |
| chrX_+_135723531 | 0.05 |
ENSMUST00000113085.2
|
Plp1
|
proteolipid protein (myelin) 1 |
| chr14_-_55114989 | 0.05 |
ENSMUST00000168622.2
ENSMUST00000177403.2 |
Ppp1r3e
|
protein phosphatase 1, regulatory subunit 3E |
| chr4_-_63414188 | 0.04 |
ENSMUST00000063650.10
ENSMUST00000102867.8 ENSMUST00000107393.8 ENSMUST00000084510.8 ENSMUST00000095038.8 ENSMUST00000119294.8 ENSMUST00000095037.2 ENSMUST00000063672.10 |
Whrn
|
whirlin |
| chr7_+_101732323 | 0.03 |
ENSMUST00000124189.3
|
Trpc2
|
transient receptor potential cation channel, subfamily C, member 2 |
| chr7_+_24584076 | 0.02 |
ENSMUST00000153451.9
ENSMUST00000108429.8 |
Rps19
|
ribosomal protein S19 |
| chr2_-_104324035 | 0.01 |
ENSMUST00000111124.8
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr4_-_95965747 | 0.01 |
ENSMUST00000097973.3
|
Cyp2j13
|
cytochrome P450, family 2, subfamily j, polypeptide 13 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.3 | 1.6 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.2 | 1.2 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.2 | 1.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.2 | 1.7 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 0.4 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.1 | 2.0 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 | 2.0 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.1 | 0.4 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.1 | 0.3 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.1 | 1.0 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.5 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 1.8 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.0 | 0.3 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.0 | 0.2 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.2 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.5 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.2 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.3 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 1.0 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.6 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.3 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.2 | 1.0 | GO:0043293 | apoptosome(GO:0043293) |
| 0.2 | 0.5 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.1 | 1.8 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.2 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 0.3 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 0.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 1.1 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.3 | 1.2 | GO:0038100 | nodal binding(GO:0038100) |
| 0.3 | 2.0 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 1.6 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.2 | 1.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.4 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 0.2 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 1.0 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.5 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.3 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 1.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 2.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.3 | GO:0001727 | lipid kinase activity(GO:0001727) diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 1.4 | GO:0004843 | cysteine-type endopeptidase activity(GO:0004197) thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.5 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.5 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 2.0 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 1.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.8 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |