Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Trp73
Z-value: 0.84
Transcription factors associated with Trp73
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Trp73
|
ENSMUSG00000029026.18 | Trp73 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Trp73 | mm39_v1_chr4_-_154181562_154181630 | 0.48 | 3.0e-03 | Click! |
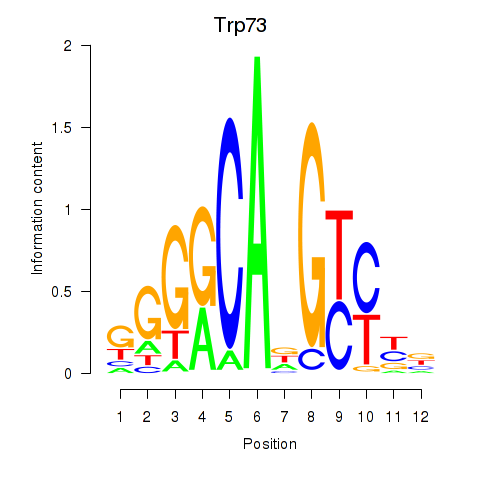
Activity profile of Trp73 motif
Sorted Z-values of Trp73 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Trp73
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_32246489 | 6.12 |
ENSMUST00000093207.4
|
Hba-a2
|
hemoglobin alpha, adult chain 2 |
| chr11_+_32233511 | 4.68 |
ENSMUST00000093209.4
|
Hba-a1
|
hemoglobin alpha, adult chain 1 |
| chr4_-_87724533 | 2.95 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr5_+_137286535 | 2.68 |
ENSMUST00000024099.11
ENSMUST00000196208.5 ENSMUST00000085934.4 |
Ache
|
acetylcholinesterase |
| chr11_-_69838971 | 1.91 |
ENSMUST00000179298.3
ENSMUST00000018710.13 ENSMUST00000135437.3 ENSMUST00000141837.9 ENSMUST00000142500.8 |
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr17_+_29309942 | 1.89 |
ENSMUST00000119901.9
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chr7_-_141023199 | 1.78 |
ENSMUST00000106005.9
|
Pidd1
|
p53 induced death domain protein 1 |
| chr3_-_129763801 | 1.76 |
ENSMUST00000029624.15
|
Mcub
|
mitochondrial calcium uniporter dominant negative beta subunit |
| chr8_+_27575611 | 1.72 |
ENSMUST00000178514.8
ENSMUST00000033876.14 |
Adgra2
|
adhesion G protein-coupled receptor A2 |
| chr16_-_16942970 | 1.70 |
ENSMUST00000093336.8
ENSMUST00000231681.2 |
2610318N02Rik
|
RIKEN cDNA 2610318N02 gene |
| chr3_-_129763638 | 1.70 |
ENSMUST00000146340.2
ENSMUST00000153506.8 |
Mcub
|
mitochondrial calcium uniporter dominant negative beta subunit |
| chr6_-_29212295 | 1.63 |
ENSMUST00000078155.12
|
Impdh1
|
inosine monophosphate dehydrogenase 1 |
| chr4_-_87724512 | 1.62 |
ENSMUST00000148059.2
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr6_-_29212239 | 1.60 |
ENSMUST00000160878.8
|
Impdh1
|
inosine monophosphate dehydrogenase 1 |
| chr11_+_97917746 | 1.51 |
ENSMUST00000017548.7
|
Rpl19
|
ribosomal protein L19 |
| chr2_+_150751475 | 1.50 |
ENSMUST00000028948.5
|
Gins1
|
GINS complex subunit 1 (Psf1 homolog) |
| chr11_+_69804714 | 1.20 |
ENSMUST00000072581.9
ENSMUST00000116358.8 |
Gps2
|
G protein pathway suppressor 2 |
| chr11_-_103235475 | 1.18 |
ENSMUST00000041385.14
|
Arhgap27
|
Rho GTPase activating protein 27 |
| chr13_-_55169100 | 1.06 |
ENSMUST00000148221.8
ENSMUST00000052949.13 |
Hk3
|
hexokinase 3 |
| chr2_-_91854844 | 1.01 |
ENSMUST00000028663.5
|
Creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chr15_-_98505508 | 1.00 |
ENSMUST00000096224.6
|
Adcy6
|
adenylate cyclase 6 |
| chr3_+_159545309 | 1.00 |
ENSMUST00000068952.10
ENSMUST00000198878.2 |
Wls
|
wntless WNT ligand secretion mediator |
| chr13_-_55169000 | 0.99 |
ENSMUST00000153665.8
|
Hk3
|
hexokinase 3 |
| chr6_-_83031358 | 0.92 |
ENSMUST00000113962.8
ENSMUST00000089645.13 ENSMUST00000113963.8 |
Htra2
|
HtrA serine peptidase 2 |
| chr7_-_97387145 | 0.92 |
ENSMUST00000084986.8
|
Aqp11
|
aquaporin 11 |
| chr7_-_115637970 | 0.79 |
ENSMUST00000166877.8
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr5_+_135698881 | 0.76 |
ENSMUST00000153500.8
|
Por
|
P450 (cytochrome) oxidoreductase |
| chr9_-_58065800 | 0.75 |
ENSMUST00000168864.4
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr5_+_64960705 | 0.74 |
ENSMUST00000165536.8
|
Klf3
|
Kruppel-like factor 3 (basic) |
| chr11_-_76359222 | 0.73 |
ENSMUST00000065028.14
|
Abr
|
active BCR-related gene |
| chr10_+_110581293 | 0.65 |
ENSMUST00000174857.8
ENSMUST00000073781.12 ENSMUST00000173471.8 ENSMUST00000173634.2 |
E2f7
|
E2F transcription factor 7 |
| chrX_-_10083157 | 0.65 |
ENSMUST00000072393.9
ENSMUST00000044598.13 ENSMUST00000073392.11 ENSMUST00000115533.8 ENSMUST00000115532.2 |
Rpgr
|
retinitis pigmentosa GTPase regulator |
| chr9_-_44624496 | 0.63 |
ENSMUST00000144251.8
ENSMUST00000156918.8 |
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr19_+_53317844 | 0.56 |
ENSMUST00000111737.3
ENSMUST00000025998.15 ENSMUST00000237837.2 |
Mxi1
|
MAX interactor 1, dimerization protein |
| chr17_-_46513499 | 0.55 |
ENSMUST00000024749.9
|
Polh
|
polymerase (DNA directed), eta (RAD 30 related) |
| chr7_+_127344942 | 0.51 |
ENSMUST00000189562.7
ENSMUST00000186116.7 |
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr4_+_108857803 | 0.51 |
ENSMUST00000065977.11
ENSMUST00000102736.9 |
Nrd1
|
nardilysin, N-arginine dibasic convertase, NRD convertase 1 |
| chr11_+_60030891 | 0.47 |
ENSMUST00000171108.8
ENSMUST00000090806.5 |
Rai1
|
retinoic acid induced 1 |
| chr19_-_47126719 | 0.46 |
ENSMUST00000140512.8
ENSMUST00000035822.2 |
Calhm2
|
calcium homeostasis modulator family member 2 |
| chr4_+_108857967 | 0.46 |
ENSMUST00000106644.9
|
Nrd1
|
nardilysin, N-arginine dibasic convertase, NRD convertase 1 |
| chr15_+_98953044 | 0.45 |
ENSMUST00000230021.2
ENSMUST00000047104.15 ENSMUST00000024249.5 |
Prph
|
peripherin |
| chr14_-_24295988 | 0.40 |
ENSMUST00000073687.13
ENSMUST00000090398.11 |
Dlg5
|
discs large MAGUK scaffold protein 5 |
| chr6_+_41095752 | 0.39 |
ENSMUST00000103269.3
|
Trbv12-2
|
T cell receptor beta, variable 12-2 |
| chr5_-_103777145 | 0.36 |
ENSMUST00000031263.2
|
Slc10a6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6 |
| chr18_-_32170012 | 0.35 |
ENSMUST00000134663.2
|
Myo7b
|
myosin VIIB |
| chr4_+_32238712 | 0.33 |
ENSMUST00000108180.9
|
Bach2
|
BTB and CNC homology, basic leucine zipper transcription factor 2 |
| chr2_-_180562718 | 0.33 |
ENSMUST00000037299.15
ENSMUST00000108876.9 |
Ythdf1
|
YTH N6-methyladenosine RNA binding protein 1 |
| chr14_+_53607470 | 0.28 |
ENSMUST00000103652.5
|
Trav14n-3
|
T cell receptor alpha variable 14N-3 |
| chr4_-_49383576 | 0.28 |
ENSMUST00000107698.8
|
Acnat2
|
acyl-coenzyme A amino acid N-acyltransferase 2 |
| chr19_+_46292759 | 0.28 |
ENSMUST00000237330.2
|
Nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
| chr4_+_100926170 | 0.26 |
ENSMUST00000106955.2
ENSMUST00000038463.15 |
Raver2
|
ribonucleoprotein, PTB-binding 2 |
| chr12_-_84923252 | 0.25 |
ENSMUST00000163189.8
ENSMUST00000110254.9 ENSMUST00000002073.13 |
Ltbp2
|
latent transforming growth factor beta binding protein 2 |
| chr11_+_75546989 | 0.25 |
ENSMUST00000136935.2
|
Myo1c
|
myosin IC |
| chr7_-_4517367 | 0.24 |
ENSMUST00000166161.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr9_-_32454157 | 0.24 |
ENSMUST00000183767.2
|
Fli1
|
Friend leukemia integration 1 |
| chr4_+_127137577 | 0.23 |
ENSMUST00000142029.2
|
Smim12
|
small integral membrane protein 12 |
| chr17_+_28910714 | 0.22 |
ENSMUST00000233250.2
|
Mapk14
|
mitogen-activated protein kinase 14 |
| chr3_+_28752050 | 0.21 |
ENSMUST00000029240.14
|
Slc2a2
|
solute carrier family 2 (facilitated glucose transporter), member 2 |
| chr14_+_33807935 | 0.20 |
ENSMUST00000022519.15
|
Anxa8
|
annexin A8 |
| chr19_+_46293210 | 0.20 |
ENSMUST00000236591.2
|
Nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
| chr7_+_140462343 | 0.19 |
ENSMUST00000163610.9
ENSMUST00000164681.8 |
Psmd13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr1_+_75483721 | 0.19 |
ENSMUST00000037330.5
|
Inha
|
inhibin alpha |
| chr7_+_140461860 | 0.18 |
ENSMUST00000026560.14
|
Psmd13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr14_-_52341426 | 0.18 |
ENSMUST00000227536.2
ENSMUST00000227195.2 ENSMUST00000228815.2 ENSMUST00000228198.2 ENSMUST00000227458.2 ENSMUST00000228232.2 ENSMUST00000227242.2 ENSMUST00000228748.2 |
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr6_-_97408367 | 0.18 |
ENSMUST00000124050.3
|
Frmd4b
|
FERM domain containing 4B |
| chr2_-_75808508 | 0.17 |
ENSMUST00000099995.5
|
Ttc30a2
|
tetratricopeptide repeat domain 30A2 |
| chr19_-_11261177 | 0.17 |
ENSMUST00000186937.7
ENSMUST00000067673.13 |
Ms4a5
|
membrane-spanning 4-domains, subfamily A, member 5 |
| chr12_+_3415107 | 0.16 |
ENSMUST00000220210.2
|
Kif3c
|
kinesin family member 3C |
| chr15_-_7427815 | 0.14 |
ENSMUST00000096494.5
|
Egflam
|
EGF-like, fibronectin type III and laminin G domains |
| chr16_-_56748424 | 0.14 |
ENSMUST00000210579.2
|
Lnp1
|
leukemia NUP98 fusion partner 1 |
| chr4_-_63414188 | 0.14 |
ENSMUST00000063650.10
ENSMUST00000102867.8 ENSMUST00000107393.8 ENSMUST00000084510.8 ENSMUST00000095038.8 ENSMUST00000119294.8 ENSMUST00000095037.2 ENSMUST00000063672.10 |
Whrn
|
whirlin |
| chr13_+_58955675 | 0.13 |
ENSMUST00000224402.2
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr2_+_119067832 | 0.12 |
ENSMUST00000028783.14
|
Spint1
|
serine protease inhibitor, Kunitz type 1 |
| chr7_+_30487322 | 0.12 |
ENSMUST00000189673.7
ENSMUST00000190990.7 ENSMUST00000189962.7 ENSMUST00000187493.7 ENSMUST00000098559.3 |
Krtdap
|
keratinocyte differentiation associated protein |
| chr1_+_55127110 | 0.12 |
ENSMUST00000075242.7
|
Hspe1
|
heat shock protein 1 (chaperonin 10) |
| chr15_-_7427759 | 0.12 |
ENSMUST00000058593.10
|
Egflam
|
EGF-like, fibronectin type III and laminin G domains |
| chr19_+_46293160 | 0.11 |
ENSMUST00000073116.13
|
Nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
| chr11_-_102120917 | 0.11 |
ENSMUST00000008999.12
|
Hdac5
|
histone deacetylase 5 |
| chr12_+_3415143 | 0.11 |
ENSMUST00000020999.7
|
Kif3c
|
kinesin family member 3C |
| chr11_+_46295547 | 0.11 |
ENSMUST00000063166.6
|
Fam71b
|
family with sequence similarity 71, member B |
| chr6_-_97464761 | 0.10 |
ENSMUST00000032146.14
|
Frmd4b
|
FERM domain containing 4B |
| chr17_+_28451674 | 0.10 |
ENSMUST00000002320.16
ENSMUST00000232879.2 ENSMUST00000166744.8 |
Ppard
|
peroxisome proliferator activator receptor delta |
| chr15_-_76259126 | 0.08 |
ENSMUST00000071119.8
|
Tssk5
|
testis-specific serine kinase 5 |
| chr11_+_98632953 | 0.08 |
ENSMUST00000153043.8
|
Thra
|
thyroid hormone receptor alpha |
| chr2_+_119068012 | 0.08 |
ENSMUST00000110817.3
|
Spint1
|
serine protease inhibitor, Kunitz type 1 |
| chr10_-_105677184 | 0.07 |
ENSMUST00000046638.10
|
Mettl25
|
methyltransferase like 25 |
| chr11_-_102120953 | 0.06 |
ENSMUST00000107150.8
ENSMUST00000156337.2 ENSMUST00000107151.9 ENSMUST00000107152.9 |
Hdac5
|
histone deacetylase 5 |
| chr11_+_69914746 | 0.06 |
ENSMUST00000124568.10
|
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr7_-_4517608 | 0.04 |
ENSMUST00000166959.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr16_+_11802445 | 0.03 |
ENSMUST00000170672.9
ENSMUST00000023138.8 |
Shisa9
|
shisa family member 9 |
| chr13_+_58956077 | 0.02 |
ENSMUST00000109838.10
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr17_+_34159633 | 0.00 |
ENSMUST00000025170.11
|
Wdr46
|
WD repeat domain 46 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0045212 | acetylcholine metabolic process(GO:0008291) neurotransmitter receptor biosynthetic process(GO:0045212) acetate ester metabolic process(GO:1900619) |
| 0.5 | 3.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.3 | 1.0 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.3 | 1.0 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.3 | 3.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.3 | 0.9 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.2 | 1.7 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.2 | 0.9 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.2 | 4.6 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 | 0.7 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.2 | 3.7 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.2 | 1.0 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.2 | 0.8 | GO:0090345 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.2 | 11.6 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.1 | 1.5 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.1 | 0.6 | GO:0002266 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.1 | 0.4 | GO:0045186 | zonula adherens assembly(GO:0045186) |
| 0.1 | 2.0 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.1 | 0.7 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.1 | 0.4 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.1 | 0.5 | GO:0006290 | pyrimidine dimer repair(GO:0006290) cellular response to UV-C(GO:0071494) |
| 0.1 | 1.9 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.1 | 0.2 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.0 | 0.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.2 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.6 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 1.0 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.2 | GO:2001184 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) stress-induced premature senescence(GO:0090400) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.0 | 0.1 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 | 1.2 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.8 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.5 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.2 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 10.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.6 | 1.9 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.5 | 1.5 | GO:0000811 | GINS complex(GO:0000811) |
| 0.5 | 3.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 2.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.6 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 1.9 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.2 | GO:0043512 | inhibin-betaglycan-ActRII complex(GO:0034673) inhibin A complex(GO:0043512) |
| 0.1 | 4.0 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 0.4 | GO:0044299 | C-fiber(GO:0044299) |
| 0.1 | 0.9 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 1.0 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:1990696 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.0 | 0.3 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 1.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.8 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.9 | 2.7 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.6 | 1.9 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.5 | 3.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.4 | 2.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.4 | 1.5 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.3 | 2.0 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 3.5 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.2 | 0.8 | GO:0016708 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.1 | 1.0 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 1.5 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 1.0 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 0.9 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 1.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.5 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.0 | 0.2 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.0 | 0.1 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 2.7 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.6 | ST GAQ PATHWAY | G alpha q Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 2.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.9 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 1.0 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 2.0 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.6 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |