Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Twist1
Z-value: 0.85
Transcription factors associated with Twist1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Twist1
|
ENSMUSG00000035799.7 | Twist1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Twist1 | mm39_v1_chr12_+_34007645_34007670 | -0.04 | 8.0e-01 | Click! |
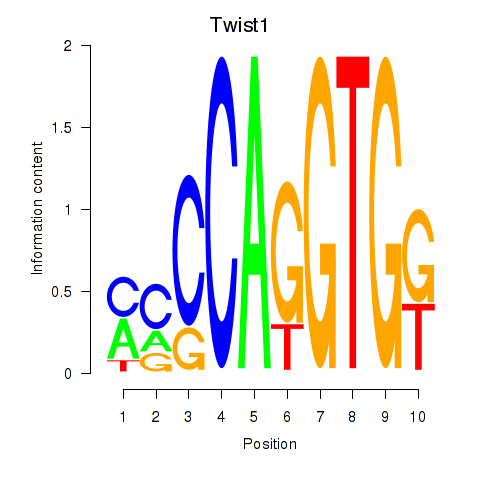
Activity profile of Twist1 motif
Sorted Z-values of Twist1 motif
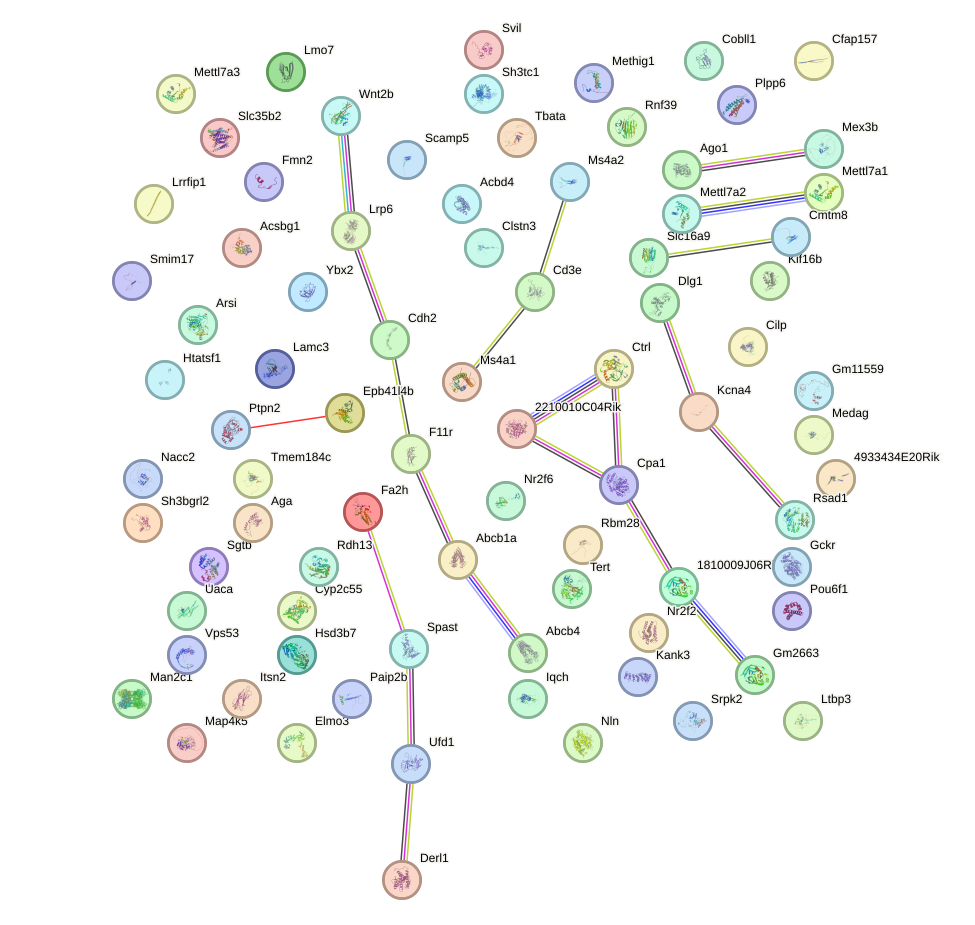
Network of associatons between targets according to the STRING database.
First level regulatory network of Twist1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_41012435 | 19.40 |
ENSMUST00000031931.6
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr6_+_30639217 | 16.72 |
ENSMUST00000031806.10
|
Cpa1
|
carboxypeptidase A1, pancreatic |
| chr8_-_106660470 | 12.46 |
ENSMUST00000034368.8
|
Ctrl
|
chymotrypsin-like |
| chr7_-_142233270 | 6.19 |
ENSMUST00000162317.2
ENSMUST00000125933.2 ENSMUST00000105931.8 ENSMUST00000105930.8 ENSMUST00000105933.8 ENSMUST00000105932.2 ENSMUST00000000220.3 |
Ins2
|
insulin II |
| chr15_+_100202642 | 1.63 |
ENSMUST00000067752.5
ENSMUST00000229588.2 |
Mettl7a1
|
methyltransferase like 7A1 |
| chr18_-_16942289 | 1.47 |
ENSMUST00000025166.14
|
Cdh2
|
cadherin 2 |
| chr6_-_40976413 | 1.30 |
ENSMUST00000166306.3
|
Gm2663
|
predicted gene 2663 |
| chr19_+_38995463 | 1.24 |
ENSMUST00000025966.5
|
Cyp2c55
|
cytochrome P450, family 2, subfamily c, polypeptide 55 |
| chr6_+_40941688 | 1.21 |
ENSMUST00000076638.7
|
1810009J06Rik
|
RIKEN cDNA 1810009J06 gene |
| chr7_+_127400016 | 1.18 |
ENSMUST00000106271.2
ENSMUST00000138432.2 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr9_+_83430363 | 1.14 |
ENSMUST00000188241.7
ENSMUST00000113215.10 |
Sh3bgrl2
|
SH3 domain binding glutamic acid-rich protein like 2 |
| chr2_+_71811526 | 1.01 |
ENSMUST00000090826.12
ENSMUST00000102698.10 |
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr5_+_8943943 | 0.98 |
ENSMUST00000196067.2
|
Abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr1_+_171265103 | 0.91 |
ENSMUST00000043839.5
|
F11r
|
F11 receptor |
| chr2_-_26012751 | 0.91 |
ENSMUST00000140993.2
ENSMUST00000028300.6 |
Nacc2
|
nucleus accumbens associated 2, BEN and BTB (POZ) domain containing |
| chr17_+_37253802 | 0.90 |
ENSMUST00000040498.12
|
Rnf39
|
ring finger protein 39 |
| chr4_-_57143437 | 0.84 |
ENSMUST00000095076.10
ENSMUST00000030142.4 |
Epb41l4b
|
erythrocyte membrane protein band 4.1 like 4b |
| chr15_-_100497863 | 0.82 |
ENSMUST00000073837.13
|
Pou6f1
|
POU domain, class 6, transcription factor 1 |
| chr16_+_31482658 | 0.78 |
ENSMUST00000115201.8
|
Dlg1
|
discs large MAGUK scaffold protein 1 |
| chr16_-_59421342 | 0.75 |
ENSMUST00000172910.3
|
Crybg3
|
beta-gamma crystallin domain containing 3 |
| chr2_+_29855572 | 0.72 |
ENSMUST00000113719.9
ENSMUST00000113717.8 ENSMUST00000113741.8 ENSMUST00000100225.9 ENSMUST00000095083.11 ENSMUST00000046257.14 |
Sptan1
|
spectrin alpha, non-erythrocytic 1 |
| chr2_-_65068917 | 0.71 |
ENSMUST00000090896.10
ENSMUST00000155082.2 |
Cobll1
|
Cobl-like 1 |
| chr7_-_70010341 | 0.64 |
ENSMUST00000032768.15
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr8_+_106032205 | 0.60 |
ENSMUST00000109375.4
ENSMUST00000212033.2 |
Elmo3
|
engulfment and cell motility 3 |
| chr17_+_74645936 | 0.58 |
ENSMUST00000224711.2
ENSMUST00000024869.8 ENSMUST00000233611.2 |
Spast
|
spastin |
| chr8_-_78337297 | 0.58 |
ENSMUST00000141202.2
ENSMUST00000152168.8 |
Tmem184c
|
transmembrane protein 184C |
| chr16_+_31482745 | 0.57 |
ENSMUST00000100001.10
ENSMUST00000064477.14 |
Dlg1
|
discs large MAGUK scaffold protein 1 |
| chr9_+_60701749 | 0.57 |
ENSMUST00000214354.2
ENSMUST00000050183.7 ENSMUST00000217656.2 |
Uaca
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr2_+_31777273 | 0.56 |
ENSMUST00000138325.8
ENSMUST00000028187.7 |
Lamc3
|
laminin gamma 3 |
| chr6_-_134543876 | 0.53 |
ENSMUST00000032322.15
ENSMUST00000126836.4 |
Lrp6
|
low density lipoprotein receptor-related protein 6 |
| chr13_+_104246245 | 0.51 |
ENSMUST00000044385.14
|
Sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr13_-_104246084 | 0.51 |
ENSMUST00000224945.2
ENSMUST00000109315.5 |
Nln
|
neurolysin (metallopeptidase M3 family) |
| chr7_-_80051455 | 0.49 |
ENSMUST00000120753.3
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr4_-_123458465 | 0.49 |
ENSMUST00000238731.2
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr12_+_4642987 | 0.48 |
ENSMUST00000062580.8
ENSMUST00000220311.2 |
Itsn2
|
intersectin 2 |
| chr9_-_44920899 | 0.47 |
ENSMUST00000102832.3
|
Cd3e
|
CD3 antigen, epsilon polypeptide |
| chr9_-_114673158 | 0.43 |
ENSMUST00000047013.4
|
Cmtm8
|
CKLF-like MARVEL transmembrane domain containing 8 |
| chr11_+_102993865 | 0.41 |
ENSMUST00000152971.2
|
Acbd4
|
acyl-Coenzyme A binding domain containing 4 |
| chr19_-_11243530 | 0.39 |
ENSMUST00000169159.3
|
Ms4a1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr10_+_70080913 | 0.38 |
ENSMUST00000046807.7
|
Slc16a9
|
solute carrier family 16 (monocarboxylic acid transporters), member 9 |
| chr13_+_73775001 | 0.38 |
ENSMUST00000022104.9
|
Tert
|
telomerase reverse transcriptase |
| chr15_+_100251030 | 0.38 |
ENSMUST00000075675.7
ENSMUST00000088142.6 ENSMUST00000176287.2 |
Methig1
Mettl7a2
|
methyltransferase hypoxia inducible domain containing 1 methyltransferase like 7A2 |
| chr9_+_57037974 | 0.37 |
ENSMUST00000160147.8
ENSMUST00000161663.8 ENSMUST00000034836.16 ENSMUST00000161182.8 |
Man2c1
|
mannosidase, alpha, class 2C, member 1 |
| chr19_+_5790918 | 0.36 |
ENSMUST00000081496.6
|
Ltbp3
|
latent transforming growth factor beta binding protein 3 |
| chr14_+_101967343 | 0.33 |
ENSMUST00000100337.10
|
Lmo7
|
LIM domain only 7 |
| chr5_+_31454939 | 0.32 |
ENSMUST00000201675.3
|
Gckr
|
glucokinase regulatory protein |
| chr18_+_4920513 | 0.31 |
ENSMUST00000126977.8
|
Svil
|
supervillin |
| chrX_+_56098908 | 0.30 |
ENSMUST00000114751.9
ENSMUST00000088652.6 |
Htatsf1
|
HIV TAT specific factor 1 |
| chr5_+_149335214 | 0.30 |
ENSMUST00000093110.12
|
Medag
|
mesenteric estrogen dependent adipogenesis |
| chr6_-_83808717 | 0.29 |
ENSMUST00000058383.9
|
Paip2b
|
poly(A) binding protein interacting protein 2B |
| chr7_+_82516491 | 0.29 |
ENSMUST00000082237.7
|
Mex3b
|
mex3 RNA binding family member B |
| chr13_-_58261406 | 0.28 |
ENSMUST00000160860.9
|
Klhl3
|
kelch-like 3 |
| chr1_+_90926443 | 0.27 |
ENSMUST00000189505.7
ENSMUST00000185531.7 ENSMUST00000068116.13 |
Lrrfip1
|
leucine rich repeat (in FLII) interacting protein 1 |
| chr2_-_142743438 | 0.26 |
ENSMUST00000230763.2
ENSMUST00000043589.8 |
Kif16b
|
kinesin family member 16B |
| chr9_+_83430395 | 0.26 |
ENSMUST00000188030.2
|
Sh3bgrl2
|
SH3 domain binding glutamic acid-rich protein like 2 |
| chr5_-_35886605 | 0.26 |
ENSMUST00000070203.14
|
Sh3tc1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr3_+_89958940 | 0.25 |
ENSMUST00000159064.8
|
4933434E20Rik
|
RIKEN cDNA 4933434E20 gene |
| chr11_+_99755302 | 0.25 |
ENSMUST00000092694.4
|
Gm11559
|
predicted gene 11559 |
| chr2_-_142743354 | 0.25 |
ENSMUST00000211861.2
|
Kif16b
|
kinesin family member 16B |
| chr7_+_24310171 | 0.24 |
ENSMUST00000206422.2
|
Phldb3
|
pleckstrin homology like domain, family B, member 3 |
| chr5_+_31454787 | 0.23 |
ENSMUST00000201166.4
ENSMUST00000072228.9 |
Gckr
|
glucokinase regulatory protein |
| chr10_+_61010983 | 0.22 |
ENSMUST00000143207.8
ENSMUST00000148181.8 ENSMUST00000151886.8 |
Tbata
|
thymus, brain and testes associated |
| chr9_+_65172455 | 0.22 |
ENSMUST00000048762.8
|
Cilp
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| chr11_+_109541400 | 0.22 |
ENSMUST00000106676.8
|
Prkar1a
|
protein kinase, cAMP dependent regulatory, type I, alpha |
| chr8_+_53964721 | 0.22 |
ENSMUST00000211424.2
ENSMUST00000033920.6 |
Aga
|
aspartylglucosaminidase |
| chr18_-_67857594 | 0.21 |
ENSMUST00000120934.8
ENSMUST00000025420.14 ENSMUST00000122412.2 |
Ptpn2
|
protein tyrosine phosphatase, non-receptor type 2 |
| chr7_-_30814652 | 0.19 |
ENSMUST00000168884.8
ENSMUST00000108102.9 |
Hpn
|
hepsin |
| chr6_-_29164981 | 0.19 |
ENSMUST00000007993.16
|
Rbm28
|
RNA binding motif protein 28 |
| chr13_+_104246259 | 0.18 |
ENSMUST00000160322.8
ENSMUST00000159574.2 |
Sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr11_+_109541747 | 0.17 |
ENSMUST00000049527.7
|
Prkar1a
|
protein kinase, cAMP dependent regulatory, type I, alpha |
| chr7_-_4448180 | 0.15 |
ENSMUST00000138798.2
|
Rdh13
|
retinol dehydrogenase 13 (all-trans and 9-cis) |
| chr11_+_69826603 | 0.15 |
ENSMUST00000018698.12
|
Ybx2
|
Y box protein 2 |
| chr15_-_57755753 | 0.15 |
ENSMUST00000022993.7
|
Derl1
|
Der1-like domain family, member 1 |
| chr15_+_100232810 | 0.14 |
ENSMUST00000075420.6
|
Mettl7a3
|
methyltransferase like 7A3 |
| chr9_-_54569128 | 0.14 |
ENSMUST00000034822.12
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr19_+_28941292 | 0.14 |
ENSMUST00000045674.4
|
Plpp6
|
phospholipid phosphatase 6 |
| chr11_+_69826719 | 0.13 |
ENSMUST00000149194.8
|
Ybx2
|
Y box protein 2 |
| chr4_-_126362372 | 0.13 |
ENSMUST00000097888.10
ENSMUST00000239399.2 |
Ago1
|
argonaute RISC catalytic subunit 1 |
| chr17_+_45874800 | 0.12 |
ENSMUST00000224905.2
ENSMUST00000226086.2 ENSMUST00000041353.7 |
Slc35b2
|
solute carrier family 35, member B2 |
| chr6_-_124410452 | 0.12 |
ENSMUST00000124998.2
ENSMUST00000238807.2 |
Clstn3
|
calsyntenin 3 |
| chr2_+_107120934 | 0.12 |
ENSMUST00000037012.3
|
Kcna4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr9_-_63509747 | 0.11 |
ENSMUST00000080527.12
ENSMUST00000042322.11 |
Iqch
|
IQ motif containing H |
| chr3_-_104869237 | 0.11 |
ENSMUST00000029429.6
|
Wnt2b
|
wingless-type MMTV integration site family, member 2B |
| chr7_-_4448631 | 0.11 |
ENSMUST00000008579.14
|
Rdh13
|
retinol dehydrogenase 13 (all-trans and 9-cis) |
| chr8_-_71834543 | 0.11 |
ENSMUST00000002466.9
|
Nr2f6
|
nuclear receptor subfamily 2, group F, member 6 |
| chr11_-_76070325 | 0.11 |
ENSMUST00000167114.8
ENSMUST00000094015.11 ENSMUST00000108419.9 ENSMUST00000170730.3 ENSMUST00000129256.2 ENSMUST00000056601.11 |
Vps53
|
VPS53 GARP complex subunit |
| chr11_+_76070483 | 0.10 |
ENSMUST00000129853.8
|
Tlcd3a
|
TLC domain containing 3A |
| chr9_-_57375269 | 0.08 |
ENSMUST00000215059.2
ENSMUST00000046587.8 ENSMUST00000214256.2 |
Scamp5
|
secretory carrier membrane protein 5 |
| chr17_+_34029484 | 0.08 |
ENSMUST00000048560.11
ENSMUST00000172649.8 ENSMUST00000173789.2 |
Kank3
|
KN motif and ankyrin repeat domains 3 |
| chr5_-_23880939 | 0.07 |
ENSMUST00000196388.5
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr7_+_6442835 | 0.07 |
ENSMUST00000168341.2
|
Olfr1344
|
olfactory receptor 1344 |
| chr1_+_174329361 | 0.06 |
ENSMUST00000030039.13
|
Fmn2
|
formin 2 |
| chr2_-_79959802 | 0.06 |
ENSMUST00000102653.8
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr18_+_61044830 | 0.06 |
ENSMUST00000040359.6
|
Arsi
|
arylsulfatase i |
| chr16_+_18630522 | 0.05 |
ENSMUST00000115578.10
|
Ufd1
|
ubiquitin recognition factor in ER-associated degradation 1 |
| chr7_-_84339045 | 0.04 |
ENSMUST00000209165.2
|
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr2_-_45007407 | 0.04 |
ENSMUST00000176438.9
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr11_-_113600346 | 0.04 |
ENSMUST00000173655.8
ENSMUST00000100248.6 |
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr2_-_32674435 | 0.04 |
ENSMUST00000102813.2
|
Cfap157
|
cilia and flagella associated protein 157 |
| chrX_-_18327610 | 0.03 |
ENSMUST00000044188.5
|
Dipk2b
|
divergent protein kinase domain 2B |
| chr17_+_75772475 | 0.03 |
ENSMUST00000095204.6
|
Rasgrp3
|
RAS, guanyl releasing protein 3 |
| chr11_-_94440025 | 0.03 |
ENSMUST00000040487.4
|
Rsad1
|
radical S-adenosyl methionine domain containing 1 |
| chr8_-_112120442 | 0.02 |
ENSMUST00000038475.9
|
Fa2h
|
fatty acid 2-hydroxylase |
| chr4_-_135221810 | 0.02 |
ENSMUST00000105856.9
|
Nipal3
|
NIPA-like domain containing 3 |
| chr12_+_102915102 | 0.02 |
ENSMUST00000101099.12
|
Unc79
|
unc-79 homolog |
| chr9_-_54568950 | 0.01 |
ENSMUST00000128624.2
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr11_-_109188892 | 0.01 |
ENSMUST00000106706.8
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr15_-_76004395 | 0.01 |
ENSMUST00000239552.1
|
EPPK1
|
epiplakin 1 |
| chr9_-_83688294 | 0.00 |
ENSMUST00000034796.14
ENSMUST00000183614.2 |
Elovl4
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4 |
| chr18_+_40390013 | 0.00 |
ENSMUST00000096572.2
ENSMUST00000236889.2 |
Kctd16
|
potassium channel tetramerisation domain containing 16 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.2 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) neuron projection maintenance(GO:1990535) |
| 0.3 | 1.3 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.3 | 1.0 | GO:1903412 | response to bile acid(GO:1903412) cellular response to bile acid(GO:1903413) |
| 0.3 | 0.9 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.2 | 1.5 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.2 | 1.0 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.2 | 0.5 | GO:0061348 | planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) |
| 0.2 | 0.6 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.1 | 0.4 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.5 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.1 | 0.4 | GO:0036363 | transforming growth factor beta activation(GO:0036363) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 1.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.1 | 1.2 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.1 | 0.6 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.5 | GO:0009750 | response to fructose(GO:0009750) |
| 0.1 | 0.5 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.1 | 0.2 | GO:1903972 | negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.1 | 0.2 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.1 | 0.9 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.6 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.5 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.4 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.6 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.3 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.6 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.4 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.0 | 0.5 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.4 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 46.5 | GO:0006508 | proteolysis(GO:0006508) |
| 0.0 | 0.0 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.6 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 0.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.5 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.1 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.0 | 0.1 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.1 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.0 | 0.3 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.2 | 5.7 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.2 | 1.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 0.5 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 0.7 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 0.6 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 1.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.9 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.5 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.9 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.5 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.0 | 2.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0036501 | UFD1-NPL4 complex(GO:0036501) |
| 0.0 | 0.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 37.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) RISC-loading complex(GO:0070578) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 16.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.3 | 1.2 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.3 | 1.2 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 0.2 | 1.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 0.5 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.2 | 1.0 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.1 | 6.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 35.2 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.1 | 0.5 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.1 | 0.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.2 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.1 | 0.5 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.4 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 1.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.4 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.9 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.4 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 1.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.5 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |