Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
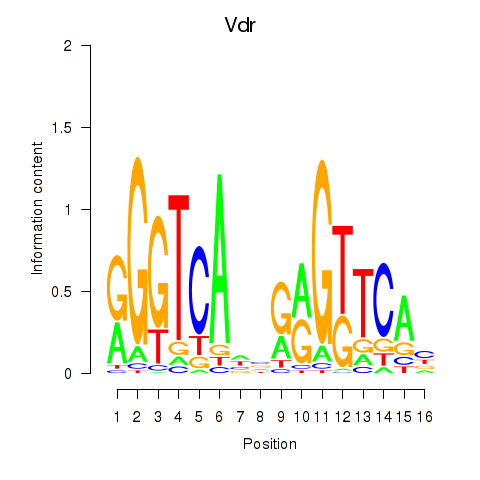
Results for Vdr
Z-value: 1.27
Transcription factors associated with Vdr
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Vdr
|
ENSMUSG00000022479.16 | Vdr |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Vdr | mm39_v1_chr15_-_97806142_97806197 | 0.21 | 2.3e-01 | Click! |
Activity profile of Vdr motif
Sorted Z-values of Vdr motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Vdr
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_110867807 | 6.44 |
ENSMUST00000197575.2
|
Ltf
|
lactotransferrin |
| chr12_+_111383864 | 5.44 |
ENSMUST00000220537.2
ENSMUST00000223050.2 ENSMUST00000072646.8 ENSMUST00000223431.2 ENSMUST00000221144.2 ENSMUST00000222437.2 |
Exoc3l4
|
exocyst complex component 3-like 4 |
| chr4_-_46413484 | 5.02 |
ENSMUST00000071096.3
|
Hemgn
|
hemogen |
| chr17_+_47816042 | 4.96 |
ENSMUST00000183044.8
ENSMUST00000037333.17 |
Ccnd3
|
cyclin D3 |
| chr10_+_127561259 | 4.83 |
ENSMUST00000026466.5
|
Tac2
|
tachykinin 2 |
| chr17_+_47816137 | 4.56 |
ENSMUST00000182935.8
ENSMUST00000182506.8 |
Ccnd3
|
cyclin D3 |
| chr17_+_47816074 | 4.46 |
ENSMUST00000183177.8
ENSMUST00000182848.8 |
Ccnd3
|
cyclin D3 |
| chr17_+_47815968 | 3.98 |
ENSMUST00000182129.8
ENSMUST00000171031.8 |
Ccnd3
|
cyclin D3 |
| chr7_-_125968653 | 3.65 |
ENSMUST00000205642.2
ENSMUST00000032997.8 ENSMUST00000206793.2 |
Lat
|
linker for activation of T cells |
| chr15_-_36598263 | 3.53 |
ENSMUST00000155116.2
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr19_-_10181243 | 3.52 |
ENSMUST00000142241.2
ENSMUST00000116542.9 ENSMUST00000025651.6 ENSMUST00000156291.2 |
Fen1
|
flap structure specific endonuclease 1 |
| chr6_+_86605146 | 3.50 |
ENSMUST00000043400.9
|
Asprv1
|
aspartic peptidase, retroviral-like 1 |
| chr7_+_142558783 | 3.29 |
ENSMUST00000009396.13
|
Tspan32
|
tetraspanin 32 |
| chr15_+_79230777 | 3.29 |
ENSMUST00000229130.2
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr17_-_33932346 | 3.27 |
ENSMUST00000173392.2
|
Marchf2
|
membrane associated ring-CH-type finger 2 |
| chr6_-_70769135 | 2.88 |
ENSMUST00000066134.6
|
Rpia
|
ribose 5-phosphate isomerase A |
| chr11_+_115790768 | 2.82 |
ENSMUST00000152171.8
|
Smim5
|
small integral membrane protein 5 |
| chr1_+_134109888 | 2.77 |
ENSMUST00000156873.8
|
Chil1
|
chitinase-like 1 |
| chr9_+_119943916 | 2.47 |
ENSMUST00000135514.2
|
Slc25a38
|
solute carrier family 25, member 38 |
| chr7_-_143056252 | 2.46 |
ENSMUST00000010904.5
|
Phlda2
|
pleckstrin homology like domain, family A, member 2 |
| chr18_+_82493237 | 2.43 |
ENSMUST00000091789.11
ENSMUST00000114676.8 |
Mbp
|
myelin basic protein |
| chr7_+_126376099 | 2.40 |
ENSMUST00000038614.12
ENSMUST00000170882.8 ENSMUST00000106359.2 ENSMUST00000106357.8 ENSMUST00000145762.8 |
Ypel3
|
yippee like 3 |
| chr4_-_41314877 | 2.39 |
ENSMUST00000030145.9
|
Dcaf12
|
DDB1 and CUL4 associated factor 12 |
| chr9_-_37464200 | 2.35 |
ENSMUST00000065668.12
|
Nrgn
|
neurogranin |
| chr5_-_148336711 | 2.34 |
ENSMUST00000048116.15
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr7_+_127347339 | 2.32 |
ENSMUST00000206893.2
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr13_-_24945844 | 2.19 |
ENSMUST00000006898.10
ENSMUST00000110382.9 |
Gmnn
|
geminin |
| chr11_+_58269862 | 2.08 |
ENSMUST00000013787.11
ENSMUST00000108826.3 |
Lypd8
|
LY6/PLAUR domain containing 8 |
| chr9_-_57169872 | 1.92 |
ENSMUST00000213199.2
ENSMUST00000034846.7 |
1700017B05Rik
|
RIKEN cDNA 1700017B05 gene |
| chr1_+_136059101 | 1.86 |
ENSMUST00000075164.11
|
Kif21b
|
kinesin family member 21B |
| chr7_+_127347308 | 1.85 |
ENSMUST00000188580.3
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr8_-_85567256 | 1.84 |
ENSMUST00000003911.13
ENSMUST00000109761.9 ENSMUST00000128035.2 |
Rad23a
|
RAD23 homolog A, nucleotide excision repair protein |
| chr7_+_126376353 | 1.83 |
ENSMUST00000106356.2
|
Ypel3
|
yippee like 3 |
| chrX_+_133486391 | 1.82 |
ENSMUST00000113211.8
|
Rpl36a
|
ribosomal protein L36A |
| chr10_+_80128248 | 1.82 |
ENSMUST00000068408.14
ENSMUST00000062674.7 |
Rps15
|
ribosomal protein S15 |
| chr9_-_57169830 | 1.81 |
ENSMUST00000215298.2
|
1700017B05Rik
|
RIKEN cDNA 1700017B05 gene |
| chr7_+_27259895 | 1.75 |
ENSMUST00000187032.2
|
2310022A10Rik
|
RIKEN cDNA 2310022A10 gene |
| chr19_-_34231600 | 1.67 |
ENSMUST00000238147.2
|
Acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr15_+_89218601 | 1.64 |
ENSMUST00000023282.9
|
Miox
|
myo-inositol oxygenase |
| chr5_-_115236354 | 1.62 |
ENSMUST00000100848.3
|
Gm10401
|
predicted gene 10401 |
| chrX_-_48877080 | 1.53 |
ENSMUST00000114893.8
|
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr19_-_8775935 | 1.52 |
ENSMUST00000096261.5
|
Polr2g
|
polymerase (RNA) II (DNA directed) polypeptide G |
| chr9_-_72399221 | 1.48 |
ENSMUST00000185151.8
ENSMUST00000085358.12 ENSMUST00000184125.8 ENSMUST00000183574.8 ENSMUST00000184831.8 |
Tex9
|
testis expressed gene 9 |
| chr12_-_113393439 | 1.46 |
ENSMUST00000103430.2
|
Ighj1
|
immunoglobulin heavy joining 1 |
| chr10_-_80184238 | 1.46 |
ENSMUST00000095446.10
ENSMUST00000105352.2 |
Adamtsl5
|
ADAMTS-like 5 |
| chr10_-_117628565 | 1.41 |
ENSMUST00000167943.8
ENSMUST00000064848.7 |
Nup107
|
nucleoporin 107 |
| chr11_-_84710276 | 1.40 |
ENSMUST00000018549.8
|
Mrm1
|
mitochondrial rRNA methyltransferase 1 |
| chr8_-_81466126 | 1.36 |
ENSMUST00000043359.9
|
Smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr7_+_12656217 | 1.33 |
ENSMUST00000108539.8
ENSMUST00000004554.14 ENSMUST00000147435.8 ENSMUST00000137329.4 |
Rps5
|
ribosomal protein S5 |
| chr7_-_141023902 | 1.33 |
ENSMUST00000026580.12
|
Pidd1
|
p53 induced death domain protein 1 |
| chr11_+_68792364 | 1.28 |
ENSMUST00000073471.13
ENSMUST00000101014.9 ENSMUST00000128952.8 ENSMUST00000167436.3 |
Rpl26
|
ribosomal protein L26 |
| chr2_+_128942900 | 1.23 |
ENSMUST00000103205.11
|
Polr1b
|
polymerase (RNA) I polypeptide B |
| chr5_-_33375509 | 1.21 |
ENSMUST00000201475.2
|
Spon2
|
spondin 2, extracellular matrix protein |
| chr19_-_8775817 | 1.20 |
ENSMUST00000235964.2
|
Polr2g
|
polymerase (RNA) II (DNA directed) polypeptide G |
| chr19_+_10181378 | 1.20 |
ENSMUST00000040372.14
|
Tmem258
|
transmembrane protein 258 |
| chr15_+_101310283 | 1.19 |
ENSMUST00000068904.9
|
Krt7
|
keratin 7 |
| chr8_+_106607507 | 1.18 |
ENSMUST00000040254.16
ENSMUST00000119261.8 |
Edc4
|
enhancer of mRNA decapping 4 |
| chr4_-_43578823 | 1.16 |
ENSMUST00000030189.14
|
Gba2
|
glucosidase beta 2 |
| chr10_+_80100812 | 1.15 |
ENSMUST00000105362.8
ENSMUST00000105361.10 |
Dazap1
|
DAZ associated protein 1 |
| chr11_+_62539657 | 1.12 |
ENSMUST00000127589.2
ENSMUST00000155759.3 |
Mmgt2
|
membrane magnesium transporter 2 |
| chr7_-_16651107 | 1.12 |
ENSMUST00000173139.2
|
Calm3
|
calmodulin 3 |
| chr11_-_62539284 | 1.12 |
ENSMUST00000057194.9
|
Lrrc75a
|
leucine rich repeat containing 75A |
| chr11_-_120520954 | 1.12 |
ENSMUST00000106180.2
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chr19_+_37184927 | 1.11 |
ENSMUST00000024078.15
ENSMUST00000112391.8 |
Marchf5
|
membrane associated ring-CH-type finger 5 |
| chr1_+_160022785 | 1.03 |
ENSMUST00000135680.8
ENSMUST00000097193.3 |
Mrps14
|
mitochondrial ribosomal protein S14 |
| chr4_+_109272828 | 1.02 |
ENSMUST00000106618.8
|
Ttc39a
|
tetratricopeptide repeat domain 39A |
| chr10_-_80742244 | 1.00 |
ENSMUST00000105332.3
|
Lmnb2
|
lamin B2 |
| chr11_-_120472022 | 0.97 |
ENSMUST00000067936.6
|
Arhgdia
|
Rho GDP dissociation inhibitor (GDI) alpha |
| chr2_+_128942919 | 0.97 |
ENSMUST00000028874.8
|
Polr1b
|
polymerase (RNA) I polypeptide B |
| chr17_+_34131696 | 0.95 |
ENSMUST00000174146.3
|
Daxx
|
Fas death domain-associated protein |
| chr15_-_97712711 | 0.92 |
ENSMUST00000121514.8
|
Hdac7
|
histone deacetylase 7 |
| chr11_+_102772030 | 0.90 |
ENSMUST00000021307.10
ENSMUST00000159834.2 |
Ccdc103
|
coiled-coil domain containing 103 |
| chr11_-_5848771 | 0.89 |
ENSMUST00000102921.4
|
Myl7
|
myosin, light polypeptide 7, regulatory |
| chr9_+_98169482 | 0.88 |
ENSMUST00000134253.8
|
Nmnat3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr15_-_102097387 | 0.88 |
ENSMUST00000230288.2
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr1_+_78286946 | 0.87 |
ENSMUST00000036172.10
|
Sgpp2
|
sphingosine-1-phosphate phosphatase 2 |
| chr2_+_25152600 | 0.87 |
ENSMUST00000114336.4
|
Tprn
|
taperin |
| chr16_-_18448454 | 0.85 |
ENSMUST00000231622.2
|
Septin5
|
septin 5 |
| chr7_+_15832383 | 0.84 |
ENSMUST00000006181.7
|
Napa
|
N-ethylmaleimide sensitive fusion protein attachment protein alpha |
| chrX_+_100576340 | 0.84 |
ENSMUST00000118878.8
ENSMUST00000101341.9 ENSMUST00000149274.9 |
Taf1
|
TATA-box binding protein associated factor 1 |
| chr11_+_3438274 | 0.82 |
ENSMUST00000064265.13
|
Pla2g3
|
phospholipase A2, group III |
| chr10_+_127871493 | 0.81 |
ENSMUST00000092048.13
|
Naca
|
nascent polypeptide-associated complex alpha polypeptide |
| chr18_+_82493284 | 0.79 |
ENSMUST00000047865.14
|
Mbp
|
myelin basic protein |
| chr8_-_71349927 | 0.79 |
ENSMUST00000212709.2
ENSMUST00000212796.2 ENSMUST00000212378.2 ENSMUST00000054220.10 ENSMUST00000212494.2 |
Rpl18a
|
ribosomal protein L18A |
| chr7_+_4463686 | 0.79 |
ENSMUST00000167298.2
ENSMUST00000171445.8 |
Eps8l1
|
EPS8-like 1 |
| chr11_-_114825089 | 0.79 |
ENSMUST00000149663.4
ENSMUST00000239005.2 ENSMUST00000106581.5 |
Cd300lb
|
CD300 molecule like family member B |
| chr19_-_5711650 | 0.79 |
ENSMUST00000236006.2
|
Sipa1
|
signal-induced proliferation associated gene 1 |
| chr10_-_89279751 | 0.78 |
ENSMUST00000219351.2
ENSMUST00000220128.2 ENSMUST00000099374.9 ENSMUST00000105298.3 |
Gas2l3
|
growth arrest-specific 2 like 3 |
| chr19_+_47167259 | 0.78 |
ENSMUST00000111808.11
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr2_-_180681079 | 0.74 |
ENSMUST00000067120.14
|
Chrna4
|
cholinergic receptor, nicotinic, alpha polypeptide 4 |
| chr11_-_51891575 | 0.74 |
ENSMUST00000109086.8
|
Ube2b
|
ubiquitin-conjugating enzyme E2B |
| chr14_+_22069877 | 0.72 |
ENSMUST00000161249.8
ENSMUST00000159777.8 ENSMUST00000162540.2 |
Lrmda
|
leucine rich melanocyte differentiation associated |
| chr18_+_44237577 | 0.72 |
ENSMUST00000239465.2
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr1_+_130645056 | 0.71 |
ENSMUST00000049813.6
|
Yod1
|
YOD1 deubiquitinase |
| chr10_-_79936987 | 0.71 |
ENSMUST00000218630.2
|
Sbno2
|
strawberry notch 2 |
| chr9_-_109531768 | 0.71 |
ENSMUST00000098359.4
|
Fbxw18
|
F-box and WD-40 domain protein 18 |
| chr7_-_44861235 | 0.71 |
ENSMUST00000210741.2
ENSMUST00000209466.2 |
Dkkl1
|
dickkopf-like 1 |
| chr12_+_17740831 | 0.70 |
ENSMUST00000071858.5
|
Hpcal1
|
hippocalcin-like 1 |
| chr14_+_33775423 | 0.70 |
ENSMUST00000058725.5
|
Antxrl
|
anthrax toxin receptor-like |
| chr10_+_127871444 | 0.69 |
ENSMUST00000073868.9
|
Naca
|
nascent polypeptide-associated complex alpha polypeptide |
| chr15_+_102391614 | 0.68 |
ENSMUST00000229432.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr19_+_34169629 | 0.67 |
ENSMUST00000239240.2
ENSMUST00000054956.15 |
Stambpl1
|
STAM binding protein like 1 |
| chr18_+_44237474 | 0.67 |
ENSMUST00000081271.7
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chrX_+_162873183 | 0.67 |
ENSMUST00000015545.10
|
Cltrn
|
collectrin, amino acid transport regulator |
| chr1_+_135710803 | 0.66 |
ENSMUST00000132795.8
|
Tnni1
|
troponin I, skeletal, slow 1 |
| chr11_-_103829040 | 0.65 |
ENSMUST00000133774.4
ENSMUST00000149642.3 |
Nsf
|
N-ethylmaleimide sensitive fusion protein |
| chr2_-_30305472 | 0.65 |
ENSMUST00000134120.2
ENSMUST00000102854.10 |
Crat
|
carnitine acetyltransferase |
| chr4_-_43578636 | 0.65 |
ENSMUST00000130443.2
|
Gba2
|
glucosidase beta 2 |
| chr9_+_53678801 | 0.64 |
ENSMUST00000048670.10
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr4_+_155953997 | 0.64 |
ENSMUST00000120794.8
ENSMUST00000030901.9 |
Ints11
|
integrator complex subunit 11 |
| chr15_-_102112657 | 0.63 |
ENSMUST00000231030.2
ENSMUST00000230687.2 ENSMUST00000229514.2 ENSMUST00000229345.2 |
Csad
|
cysteine sulfinic acid decarboxylase |
| chr8_+_124750133 | 0.61 |
ENSMUST00000034457.9
|
Urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr5_-_121641461 | 0.60 |
ENSMUST00000079368.5
|
Adam1b
|
a disintegrin and metallopeptidase domain 1b |
| chr5_-_124233812 | 0.59 |
ENSMUST00000031354.11
|
Abcb9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr5_-_120726721 | 0.59 |
ENSMUST00000046426.10
|
Tpcn1
|
two pore channel 1 |
| chr4_+_13784749 | 0.58 |
ENSMUST00000098256.4
|
Runx1t1
|
RUNX1 translocation partner 1 |
| chr11_-_78642480 | 0.58 |
ENSMUST00000059468.6
|
Ccnq
|
cyclin Q |
| chr9_+_53757448 | 0.56 |
ENSMUST00000048485.7
|
Sln
|
sarcolipin |
| chr14_+_75373915 | 0.56 |
ENSMUST00000122840.8
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr11_+_4014841 | 0.55 |
ENSMUST00000068322.7
|
Sec14l3
|
SEC14-like lipid binding 3 |
| chr16_+_91526289 | 0.54 |
ENSMUST00000113993.8
|
Itsn1
|
intersectin 1 (SH3 domain protein 1A) |
| chr19_-_30526916 | 0.53 |
ENSMUST00000025803.9
|
Dkk1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr14_-_20398108 | 0.51 |
ENSMUST00000224412.2
ENSMUST00000022344.4 |
Ecd
|
ecdysoneless cell cycle regulator |
| chr4_+_62883796 | 0.51 |
ENSMUST00000030043.13
ENSMUST00000107415.8 ENSMUST00000064814.6 |
Zfp618
|
zinc finger protein 618 |
| chr1_+_75498162 | 0.51 |
ENSMUST00000027414.16
ENSMUST00000113553.2 |
Stk11ip
|
serine/threonine kinase 11 interacting protein |
| chr6_+_83719966 | 0.50 |
ENSMUST00000006431.8
|
Atp6v1b1
|
ATPase, H+ transporting, lysosomal V1 subunit B1 |
| chr10_-_79938487 | 0.49 |
ENSMUST00000042771.8
|
Sbno2
|
strawberry notch 2 |
| chr4_-_118037143 | 0.49 |
ENSMUST00000050288.9
ENSMUST00000106403.8 |
Kdm4a
|
lysine (K)-specific demethylase 4A |
| chr5_+_101912939 | 0.48 |
ENSMUST00000031273.9
|
Cds1
|
CDP-diacylglycerol synthase 1 |
| chr3_+_116761916 | 0.48 |
ENSMUST00000117592.2
|
4930455H04Rik
|
RIKEN cDNA 4930455H04 gene |
| chr5_+_122988111 | 0.48 |
ENSMUST00000031434.8
ENSMUST00000198602.2 |
Rnf34
|
ring finger protein 34 |
| chr11_-_6576030 | 0.47 |
ENSMUST00000000394.14
ENSMUST00000189268.7 ENSMUST00000136682.8 |
Tbrg4
|
transforming growth factor beta regulated gene 4 |
| chr13_-_95581335 | 0.47 |
ENSMUST00000045583.9
|
Crhbp
|
corticotropin releasing hormone binding protein |
| chr8_-_123187406 | 0.47 |
ENSMUST00000006762.7
|
Snai3
|
snail family zinc finger 3 |
| chr4_+_41348996 | 0.45 |
ENSMUST00000108060.10
ENSMUST00000072866.12 |
Ubap1
|
ubiquitin-associated protein 1 |
| chr15_-_77854988 | 0.44 |
ENSMUST00000100484.6
|
Eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr10_+_126914755 | 0.44 |
ENSMUST00000039259.7
ENSMUST00000217941.2 |
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr6_+_83720022 | 0.44 |
ENSMUST00000206911.2
ENSMUST00000205763.2 |
Atp6v1b1
|
ATPase, H+ transporting, lysosomal V1 subunit B1 |
| chr9_-_15023396 | 0.43 |
ENSMUST00000159985.2
|
Hephl1
|
hephaestin-like 1 |
| chr9_+_57411279 | 0.43 |
ENSMUST00000080514.9
|
Rpp25
|
ribonuclease P/MRP 25 subunit |
| chr1_+_172784335 | 0.43 |
ENSMUST00000038432.7
|
Olfr16
|
olfactory receptor 16 |
| chr11_+_58854616 | 0.43 |
ENSMUST00000075141.7
|
Trim17
|
tripartite motif-containing 17 |
| chr17_-_25155868 | 0.42 |
ENSMUST00000115228.9
ENSMUST00000117509.8 ENSMUST00000121723.8 ENSMUST00000119115.8 ENSMUST00000121787.8 ENSMUST00000088345.12 ENSMUST00000120035.8 ENSMUST00000115229.10 ENSMUST00000178969.8 |
Mapk8ip3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr1_+_173248104 | 0.42 |
ENSMUST00000173023.2
|
Aim2
|
absent in melanoma 2 |
| chr8_+_4303067 | 0.42 |
ENSMUST00000011981.5
ENSMUST00000208316.2 |
Snapc2
|
small nuclear RNA activating complex, polypeptide 2 |
| chr1_+_57884693 | 0.42 |
ENSMUST00000169772.3
|
Spats2l
|
spermatogenesis associated, serine-rich 2-like |
| chr9_+_110595224 | 0.41 |
ENSMUST00000136695.3
|
Myl3
|
myosin, light polypeptide 3 |
| chr2_-_180680868 | 0.41 |
ENSMUST00000108851.8
|
Chrna4
|
cholinergic receptor, nicotinic, alpha polypeptide 4 |
| chr4_+_103000248 | 0.41 |
ENSMUST00000106855.2
|
Mier1
|
MEIR1 treanscription regulator |
| chrX_-_52358663 | 0.41 |
ENSMUST00000114841.2
ENSMUST00000071023.12 |
Fam122b
|
family with sequence similarity 122, member B |
| chr3_+_95067759 | 0.41 |
ENSMUST00000131742.8
ENSMUST00000090823.8 ENSMUST00000090821.10 |
Sema6c
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr11_+_83189831 | 0.41 |
ENSMUST00000176944.8
|
Ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr15_-_94302139 | 0.40 |
ENSMUST00000035342.11
ENSMUST00000155907.2 |
Adamts20
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 20 |
| chr1_+_151446893 | 0.40 |
ENSMUST00000134499.8
|
Niban1
|
niban apoptosis regulator 1 |
| chr11_+_68477812 | 0.40 |
ENSMUST00000154294.8
ENSMUST00000063006.12 |
Ccdc42
|
coiled-coil domain containing 42 |
| chr2_-_30305313 | 0.39 |
ENSMUST00000132981.9
ENSMUST00000129494.2 |
Crat
|
carnitine acetyltransferase |
| chr8_+_123920682 | 0.39 |
ENSMUST00000212409.2
|
Dpep1
|
dipeptidase 1 |
| chr13_-_12121831 | 0.39 |
ENSMUST00000021750.15
ENSMUST00000170156.3 ENSMUST00000220597.2 |
Ryr2
|
ryanodine receptor 2, cardiac |
| chr15_-_64886753 | 0.38 |
ENSMUST00000110100.3
|
Gm21961
|
predicted gene, 21961 |
| chr3_-_108133914 | 0.38 |
ENSMUST00000141387.4
|
Sypl2
|
synaptophysin-like 2 |
| chr17_-_24926589 | 0.38 |
ENSMUST00000126319.8
|
Tbl3
|
transducin (beta)-like 3 |
| chr2_+_29533799 | 0.37 |
ENSMUST00000238899.2
|
Rapgef1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr5_+_125080504 | 0.37 |
ENSMUST00000197746.2
|
Rflna
|
refilin A |
| chr3_-_107240989 | 0.36 |
ENSMUST00000061772.11
|
Rbm15
|
RNA binding motif protein 15 |
| chr4_+_156300325 | 0.36 |
ENSMUST00000105572.3
|
Perm1
|
PPARGC1 and ESRR induced regulator, muscle 1 |
| chr19_+_47167554 | 0.36 |
ENSMUST00000235290.2
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr15_-_77854711 | 0.35 |
ENSMUST00000230419.2
|
Eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr15_-_37008011 | 0.35 |
ENSMUST00000226671.2
|
Zfp706
|
zinc finger protein 706 |
| chr17_+_34148485 | 0.34 |
ENSMUST00000047503.16
|
Rgl2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr15_+_78926085 | 0.34 |
ENSMUST00000058004.4
|
Galr3
|
galanin receptor 3 |
| chr11_-_51891259 | 0.34 |
ENSMUST00000020657.13
|
Ube2b
|
ubiquitin-conjugating enzyme E2B |
| chr12_-_111638722 | 0.34 |
ENSMUST00000001304.9
|
Ckb
|
creatine kinase, brain |
| chr11_+_75546671 | 0.34 |
ENSMUST00000108431.3
|
Myo1c
|
myosin IC |
| chr11_+_62539479 | 0.33 |
ENSMUST00000062860.5
|
Mmgt2
|
membrane magnesium transporter 2 |
| chr4_-_118037240 | 0.33 |
ENSMUST00000106406.9
|
Kdm4a
|
lysine (K)-specific demethylase 4A |
| chr7_-_134100659 | 0.33 |
ENSMUST00000238294.2
|
D7Ertd443e
|
DNA segment, Chr 7, ERATO Doi 443, expressed |
| chr3_+_3573084 | 0.33 |
ENSMUST00000108393.8
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr8_+_123920233 | 0.32 |
ENSMUST00000212773.2
|
Dpep1
|
dipeptidase 1 |
| chr7_-_141927471 | 0.32 |
ENSMUST00000105988.2
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr3_-_144525255 | 0.32 |
ENSMUST00000029929.12
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr10_-_81463091 | 0.32 |
ENSMUST00000143424.2
ENSMUST00000119324.8 |
Sirt6
|
sirtuin 6 |
| chr7_-_79443536 | 0.32 |
ENSMUST00000032760.6
|
Mesp1
|
mesoderm posterior 1 |
| chr7_+_5083212 | 0.32 |
ENSMUST00000098845.10
ENSMUST00000146317.8 ENSMUST00000153169.2 ENSMUST00000045277.7 |
Epn1
|
epsin 1 |
| chr1_-_135934080 | 0.32 |
ENSMUST00000166193.9
|
Igfn1
|
immunoglobulin-like and fibronectin type III domain containing 1 |
| chr4_+_126042250 | 0.32 |
ENSMUST00000106150.3
|
Eva1b
|
eva-1 homolog B (C. elegans) |
| chr6_+_127538268 | 0.32 |
ENSMUST00000212051.2
|
Cracr2a
|
calcium release activated channel regulator 2A |
| chr17_+_47815998 | 0.32 |
ENSMUST00000183210.2
|
Ccnd3
|
cyclin D3 |
| chr4_+_133795740 | 0.31 |
ENSMUST00000121391.8
|
Crybg2
|
crystallin beta-gamma domain containing 2 |
| chr16_+_26281885 | 0.31 |
ENSMUST00000161053.8
ENSMUST00000115302.2 |
Cldn16
|
claudin 16 |
| chr10_+_7668655 | 0.31 |
ENSMUST00000015901.11
|
Ppil4
|
peptidylprolyl isomerase (cyclophilin)-like 4 |
| chr2_-_25436884 | 0.31 |
ENSMUST00000114234.2
|
Traf2
|
TNF receptor-associated factor 2 |
| chr7_-_25374472 | 0.31 |
ENSMUST00000108404.8
ENSMUST00000108405.2 ENSMUST00000079439.10 |
Tmem91
|
transmembrane protein 91 |
| chr14_-_51308899 | 0.30 |
ENSMUST00000048478.6
|
Olfr750
|
olfactory receptor 750 |
| chr3_+_89979948 | 0.30 |
ENSMUST00000121503.8
ENSMUST00000119570.8 |
Tpm3
|
tropomyosin 3, gamma |
| chrX_+_158771429 | 0.29 |
ENSMUST00000033665.9
|
Map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr6_-_57827328 | 0.28 |
ENSMUST00000203310.3
ENSMUST00000203488.3 |
Vmn1r21
|
vomeronasal 1 receptor 21 |
| chr8_-_83129134 | 0.28 |
ENSMUST00000209363.2
|
Il15
|
interleukin 15 |
| chr10_+_128583734 | 0.28 |
ENSMUST00000163377.10
|
Pym1
|
PYM homolog 1, exon junction complex associated factor |
| chr15_-_79326001 | 0.28 |
ENSMUST00000122044.8
ENSMUST00000135519.2 |
Csnk1e
|
casein kinase 1, epsilon |
| chr4_-_129015493 | 0.28 |
ENSMUST00000135763.2
ENSMUST00000149763.3 ENSMUST00000164649.8 |
Hpca
|
hippocalcin |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.4 | GO:0051673 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.8 | 2.5 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.8 | 3.2 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.7 | 2.9 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.7 | 18.3 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.6 | 2.5 | GO:0036233 | glycine import(GO:0036233) |
| 0.6 | 4.8 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.6 | 3.5 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.5 | 2.2 | GO:0017126 | nucleologenesis(GO:0017126) |
| 0.5 | 3.5 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.5 | 1.4 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.4 | 4.1 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.4 | 1.2 | GO:1903048 | regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 0.4 | 1.8 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.4 | 1.1 | GO:0033128 | positive regulation of reciprocal meiotic recombination(GO:0010845) negative regulation of histone phosphorylation(GO:0033128) |
| 0.3 | 1.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.3 | 5.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.3 | 1.3 | GO:1902164 | positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.3 | 1.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.3 | 1.5 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.3 | 1.2 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.3 | 3.3 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.3 | 0.9 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.3 | 1.4 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.3 | 0.8 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.3 | 1.5 | GO:1901228 | positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.2 | 0.7 | GO:1990168 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.2 | 1.7 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.2 | 0.6 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.2 | 0.8 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) cap-dependent translational initiation(GO:0002191) |
| 0.2 | 3.6 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.2 | 0.9 | GO:0072737 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.2 | 0.4 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.2 | 1.8 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 | 1.5 | GO:0015675 | nickel cation transport(GO:0015675) |
| 0.2 | 0.7 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.2 | 1.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.2 | 1.3 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.2 | 0.8 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.2 | 2.7 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 0.9 | GO:0034627 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 3.1 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.4 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 0.1 | 1.6 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.5 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 2.0 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 0.3 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 0.5 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.3 | GO:0031283 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.1 | 0.6 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 0.6 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.1 | 1.3 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 | 1.0 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.3 | GO:0060376 | positive regulation of mast cell differentiation(GO:0060376) |
| 0.1 | 0.8 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 0.8 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 0.1 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.1 | 2.4 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.1 | 0.4 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 4.3 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.9 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.2 | GO:0034147 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.0 | 0.8 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.3 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.7 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 1.8 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.3 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.3 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.4 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.1 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.0 | 0.4 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of neuroepithelial cell differentiation(GO:1902913) |
| 0.0 | 0.4 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.2 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.7 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.2 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.0 | 0.2 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 1.1 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.4 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.3 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 0.2 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.3 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.6 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.0 | 0.7 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.2 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) extrathymic T cell selection(GO:0045062) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.2 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.3 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 2.2 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 2.0 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.1 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) positive regulation of establishment of Sertoli cell barrier(GO:1904446) negative regulation of type B pancreatic cell development(GO:2000077) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.3 | GO:0061034 | succinyl-CoA metabolic process(GO:0006104) olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.6 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.6 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.8 | GO:0002446 | neutrophil mediated immunity(GO:0002446) |
| 0.0 | 0.3 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.4 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.2 | GO:1904668 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 1.0 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.5 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.4 | GO:0038180 | Rap protein signal transduction(GO:0032486) nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.0 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.0 | 0.1 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.3 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.0 | 0.3 | GO:0070633 | transepithelial transport(GO:0070633) |
| 0.0 | 0.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.4 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.7 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.7 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.1 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.0 | 0.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.5 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) |
| 0.0 | 0.1 | GO:0014719 | skeletal muscle satellite cell activation(GO:0014719) |
| 0.0 | 0.2 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.0 | 0.8 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.9 | GO:0007588 | excretion(GO:0007588) |
| 0.0 | 0.2 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 1.1 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.4 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.7 | 3.3 | GO:0070442 | integrin alphaIIb-beta3 complex(GO:0070442) |
| 0.5 | 1.5 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.4 | 2.8 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.3 | 18.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 5.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 1.7 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.2 | 2.6 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.2 | 2.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.2 | 2.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 3.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.8 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 1.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 1.4 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 1.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.4 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 1.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 1.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.5 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 4.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.8 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.4 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.5 | GO:0098888 | extrinsic component of presynaptic membrane(GO:0098888) |
| 0.1 | 0.3 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.3 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.1 | 2.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 0.5 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 1.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 0.4 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 4.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 3.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 3.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.2 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 1.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.3 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.9 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 1.0 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 1.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.4 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.8 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.9 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 3.0 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.8 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.4 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 1.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.4 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 1.8 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.5 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.6 | 1.8 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.5 | 1.4 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 0.3 | 18.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.3 | 1.2 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.3 | 1.2 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 0.3 | 2.9 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.3 | 0.8 | GO:0001132 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.3 | 1.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.3 | 0.8 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.2 | 7.7 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.2 | 2.8 | GO:0008061 | chitin binding(GO:0008061) |
| 0.2 | 0.9 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 1.5 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.2 | 3.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 1.5 | GO:0015099 | nickel cation transmembrane transporter activity(GO:0015099) |
| 0.2 | 2.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.2 | 1.5 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 2.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.2 | 0.8 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.2 | 0.5 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.6 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.1 | 0.9 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 1.6 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 3.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.6 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 3.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 0.3 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.1 | 1.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.3 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.7 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.1 | 0.4 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.1 | 2.7 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.1 | 0.8 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 2.4 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 1.8 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 0.3 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 4.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.5 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 1.2 | GO:0042166 | acetylcholine-gated cation channel activity(GO:0022848) acetylcholine binding(GO:0042166) |
| 0.1 | 1.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.3 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.7 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.4 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.3 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.0 | 0.7 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 1.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.4 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 1.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.3 | GO:0005047 | signal recognition particle binding(GO:0005047) endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 1.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.8 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.9 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.7 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.4 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 5.5 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.8 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.5 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.4 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 4.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.5 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 1.8 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.4 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.0 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.6 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 18.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 3.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.3 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.4 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.5 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 1.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.9 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.2 | ST G ALPHA I PATHWAY | G alpha i Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 18.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.3 | 3.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.2 | 6.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 2.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 2.7 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 2.2 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 3.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 3.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 1.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 3.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 3.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 3.9 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.1 | 1.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 1.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.4 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 4.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.9 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 1.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.8 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.4 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.5 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |