Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Zbtb7c
Z-value: 0.72
Transcription factors associated with Zbtb7c
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb7c
|
ENSMUSG00000044646.16 | Zbtb7c |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb7c | mm39_v1_chr18_+_76192529_76192571 | 0.57 | 2.9e-04 | Click! |
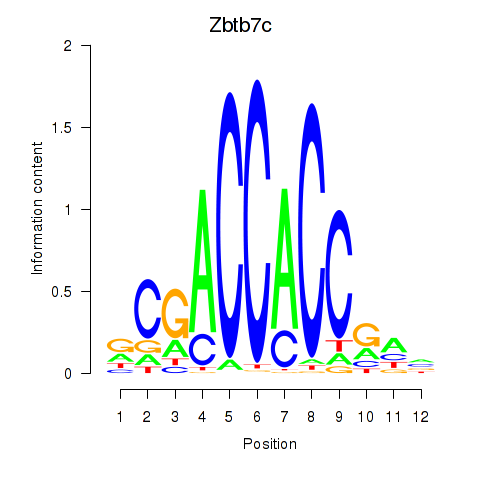
Activity profile of Zbtb7c motif
Sorted Z-values of Zbtb7c motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb7c
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_76756772 | 5.16 |
ENSMUST00000166101.2
|
Sptb
|
spectrin beta, erythrocytic |
| chr11_+_53410697 | 4.18 |
ENSMUST00000120878.9
ENSMUST00000147912.2 |
Septin8
|
septin 8 |
| chr2_+_157401998 | 3.69 |
ENSMUST00000153739.9
ENSMUST00000173595.2 ENSMUST00000109526.2 ENSMUST00000173839.2 ENSMUST00000173041.8 ENSMUST00000173793.8 ENSMUST00000172487.2 ENSMUST00000088484.6 |
Nnat
|
neuronatin |
| chr4_-_43499608 | 3.21 |
ENSMUST00000136005.3
ENSMUST00000054538.13 |
Arhgef39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr11_+_53410552 | 3.21 |
ENSMUST00000108987.8
ENSMUST00000121334.8 ENSMUST00000117061.8 |
Septin8
|
septin 8 |
| chr19_-_5323092 | 2.67 |
ENSMUST00000237463.2
ENSMUST00000025786.9 |
Pacs1
|
phosphofurin acidic cluster sorting protein 1 |
| chr10_-_93425553 | 2.17 |
ENSMUST00000020203.7
|
Snrpf
|
small nuclear ribonucleoprotein polypeptide F |
| chr2_+_72306503 | 2.05 |
ENSMUST00000102691.11
ENSMUST00000157019.2 |
Cdca7
|
cell division cycle associated 7 |
| chr15_+_76131020 | 2.00 |
ENSMUST00000229380.2
|
Grina
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr11_-_115405200 | 1.93 |
ENSMUST00000021083.7
|
Jpt1
|
Jupiter microtubule associated homolog 1 |
| chr15_+_78783867 | 1.89 |
ENSMUST00000134703.8
ENSMUST00000061239.14 ENSMUST00000109698.9 |
Gm49510
Sh3bp1
|
predicted gene, 49510 SH3-domain binding protein 1 |
| chr2_+_84867554 | 1.77 |
ENSMUST00000077798.13
|
Ssrp1
|
structure specific recognition protein 1 |
| chr18_-_35795233 | 1.72 |
ENSMUST00000025209.12
ENSMUST00000096573.4 |
Spata24
|
spermatogenesis associated 24 |
| chr11_+_115455260 | 1.63 |
ENSMUST00000021085.11
|
Nup85
|
nucleoporin 85 |
| chr15_+_78784043 | 1.63 |
ENSMUST00000001226.11
|
Sh3bp1
|
SH3-domain binding protein 1 |
| chr6_+_142702403 | 1.58 |
ENSMUST00000032419.9
|
Cmas
|
cytidine monophospho-N-acetylneuraminic acid synthetase |
| chr2_+_84867783 | 1.55 |
ENSMUST00000168266.8
ENSMUST00000130729.3 |
Ssrp1
|
structure specific recognition protein 1 |
| chr10_+_79824418 | 1.54 |
ENSMUST00000004784.11
ENSMUST00000105374.2 |
Cnn2
|
calponin 2 |
| chr19_-_41836514 | 1.38 |
ENSMUST00000059231.4
|
Frat2
|
frequently rearranged in advanced T cell lymphomas 2 |
| chr1_+_172328768 | 1.36 |
ENSMUST00000111228.2
|
Tagln2
|
transgelin 2 |
| chr18_-_35795175 | 1.31 |
ENSMUST00000236574.2
ENSMUST00000236971.2 |
Spata24
|
spermatogenesis associated 24 |
| chr3_+_87813624 | 1.29 |
ENSMUST00000005017.15
|
Hdgf
|
heparin binding growth factor |
| chr11_-_100098333 | 1.27 |
ENSMUST00000007272.8
|
Krt14
|
keratin 14 |
| chr13_-_55661240 | 1.27 |
ENSMUST00000069929.13
ENSMUST00000069968.13 ENSMUST00000131306.8 ENSMUST00000046246.13 |
Pdlim7
|
PDZ and LIM domain 7 |
| chr11_-_100026754 | 1.23 |
ENSMUST00000107411.3
|
Krt15
|
keratin 15 |
| chr10_-_80382611 | 1.22 |
ENSMUST00000183233.9
ENSMUST00000182604.8 |
Rexo1
|
REX1, RNA exonuclease 1 |
| chr16_+_48814548 | 1.19 |
ENSMUST00000117994.8
ENSMUST00000048374.6 |
Cip2a
|
cell proliferation regulating inhibitor of protein phosphatase 2A |
| chr11_-_62539284 | 1.11 |
ENSMUST00000057194.9
|
Lrrc75a
|
leucine rich repeat containing 75A |
| chr15_+_76130947 | 1.10 |
ENSMUST00000229772.2
ENSMUST00000230347.2 ENSMUST00000023225.8 |
Grina
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chrX_-_141749704 | 1.08 |
ENSMUST00000041317.3
|
Ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr8_+_72993862 | 1.01 |
ENSMUST00000003117.15
ENSMUST00000212841.2 |
Ap1m1
|
adaptor-related protein complex AP-1, mu subunit 1 |
| chr11_+_69792642 | 1.00 |
ENSMUST00000177138.8
ENSMUST00000108617.10 ENSMUST00000177476.8 ENSMUST00000061837.11 |
Neurl4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr15_-_38518406 | 0.99 |
ENSMUST00000151319.8
|
Azin1
|
antizyme inhibitor 1 |
| chr1_+_135945705 | 0.96 |
ENSMUST00000063719.15
|
Tmem9
|
transmembrane protein 9 |
| chr11_-_102209767 | 0.89 |
ENSMUST00000174302.8
ENSMUST00000178839.8 ENSMUST00000006754.14 |
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr1_+_135945798 | 0.88 |
ENSMUST00000117950.2
|
Tmem9
|
transmembrane protein 9 |
| chr10_-_79710067 | 0.83 |
ENSMUST00000166023.2
ENSMUST00000167707.2 ENSMUST00000165601.8 |
Plppr3
|
phospholipid phosphatase related 3 |
| chr6_+_135174975 | 0.80 |
ENSMUST00000111915.8
ENSMUST00000111916.2 |
Fam234b
|
family with sequence similarity 234, member B |
| chr9_+_66257747 | 0.77 |
ENSMUST00000042824.13
|
Herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr18_+_34910064 | 0.76 |
ENSMUST00000043775.9
ENSMUST00000224715.2 |
Kdm3b
|
KDM3B lysine (K)-specific demethylase 3B |
| chr5_+_33176160 | 0.76 |
ENSMUST00000019109.8
|
Ywhah
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide |
| chr8_+_33876353 | 0.74 |
ENSMUST00000070340.6
ENSMUST00000078058.5 |
Purg
|
purine-rich element binding protein G |
| chr8_+_72994152 | 0.73 |
ENSMUST00000126885.2
|
Ap1m1
|
adaptor-related protein complex AP-1, mu subunit 1 |
| chr15_-_38518458 | 0.71 |
ENSMUST00000127848.2
|
Azin1
|
antizyme inhibitor 1 |
| chr12_+_31315270 | 0.70 |
ENSMUST00000002979.16
ENSMUST00000239496.2 ENSMUST00000170495.3 |
Lamb1
|
laminin B1 |
| chr14_-_54923517 | 0.70 |
ENSMUST00000125265.2
|
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr2_+_158636727 | 0.70 |
ENSMUST00000029186.14
ENSMUST00000109478.9 ENSMUST00000156893.2 |
Dhx35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr7_-_15781838 | 0.68 |
ENSMUST00000210781.2
|
Bicra
|
BRD4 interacting chromatin remodeling complex associated protein |
| chr11_+_79980210 | 0.67 |
ENSMUST00000017694.7
ENSMUST00000108239.7 |
Atad5
|
ATPase family, AAA domain containing 5 |
| chr8_+_72993913 | 0.65 |
ENSMUST00000145213.8
|
Ap1m1
|
adaptor-related protein complex AP-1, mu subunit 1 |
| chr16_+_4867876 | 0.65 |
ENSMUST00000230703.2
ENSMUST00000052449.6 |
Ubn1
|
ubinuclein 1 |
| chr5_-_148865429 | 0.63 |
ENSMUST00000149169.3
ENSMUST00000047257.15 |
Katnal1
|
katanin p60 subunit A-like 1 |
| chr6_+_135175031 | 0.63 |
ENSMUST00000130612.2
|
Fam234b
|
family with sequence similarity 234, member B |
| chr15_+_81469538 | 0.60 |
ENSMUST00000068387.11
|
Ep300
|
E1A binding protein p300 |
| chr19_-_46306506 | 0.55 |
ENSMUST00000224556.2
|
Psd
|
pleckstrin and Sec7 domain containing |
| chr17_+_35113490 | 0.54 |
ENSMUST00000052778.10
|
Zbtb12
|
zinc finger and BTB domain containing 12 |
| chr12_+_31315227 | 0.54 |
ENSMUST00000169088.8
|
Lamb1
|
laminin B1 |
| chr7_+_35285657 | 0.54 |
ENSMUST00000040844.16
ENSMUST00000188906.7 ENSMUST00000186245.7 ENSMUST00000190503.7 |
Ankrd27
|
ankyrin repeat domain 27 (VPS9 domain) |
| chr5_-_134643805 | 0.53 |
ENSMUST00000202085.2
ENSMUST00000036362.13 ENSMUST00000077636.8 |
Lat2
|
linker for activation of T cells family, member 2 |
| chr11_-_102338473 | 0.53 |
ENSMUST00000049057.5
|
Fam171a2
|
family with sequence similarity 171, member A2 |
| chr3_-_96634769 | 0.52 |
ENSMUST00000141377.8
ENSMUST00000125183.2 |
Polr3c
|
polymerase (RNA) III (DNA directed) polypeptide C |
| chr11_-_70120503 | 0.52 |
ENSMUST00000153449.2
ENSMUST00000000326.12 |
Bcl6b
|
B cell CLL/lymphoma 6, member B |
| chr14_-_54924062 | 0.51 |
ENSMUST00000147714.8
|
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr8_+_108162985 | 0.51 |
ENSMUST00000166615.3
ENSMUST00000213097.2 ENSMUST00000212205.2 |
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chrX_+_151016224 | 0.46 |
ENSMUST00000112588.9
ENSMUST00000082177.13 |
Kdm5c
|
lysine (K)-specific demethylase 5C |
| chr5_-_137784912 | 0.42 |
ENSMUST00000031740.16
|
Mepce
|
methylphosphate capping enzyme |
| chr19_+_40600836 | 0.40 |
ENSMUST00000134063.8
|
Entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr8_+_96551974 | 0.40 |
ENSMUST00000074053.6
|
Sap18b
|
Sin3-associated polypeptide 18B |
| chr7_+_45289391 | 0.40 |
ENSMUST00000148532.4
|
Mamstr
|
MEF2 activating motif and SAP domain containing transcriptional regulator |
| chr11_+_75422516 | 0.36 |
ENSMUST00000149727.8
ENSMUST00000108433.8 ENSMUST00000042561.14 ENSMUST00000143035.8 |
Slc43a2
|
solute carrier family 43, member 2 |
| chr15_-_101602734 | 0.35 |
ENSMUST00000023788.8
|
Krt6a
|
keratin 6A |
| chr14_-_55231998 | 0.34 |
ENSMUST00000227518.2
ENSMUST00000226424.2 ENSMUST00000153783.2 ENSMUST00000102803.11 ENSMUST00000168485.8 |
Myh7
|
myosin, heavy polypeptide 7, cardiac muscle, beta |
| chr3_-_96634752 | 0.32 |
ENSMUST00000154679.8
|
Polr3c
|
polymerase (RNA) III (DNA directed) polypeptide C |
| chr8_+_4216556 | 0.31 |
ENSMUST00000239400.2
ENSMUST00000177053.8 ENSMUST00000176149.9 ENSMUST00000176072.9 ENSMUST00000176825.3 |
Evi5l
|
ecotropic viral integration site 5 like |
| chr14_+_119092107 | 0.30 |
ENSMUST00000100314.4
|
Cldn10
|
claudin 10 |
| chr10_-_79710044 | 0.30 |
ENSMUST00000167897.8
|
Plppr3
|
phospholipid phosphatase related 3 |
| chr5_-_137784943 | 0.29 |
ENSMUST00000132726.2
|
Mepce
|
methylphosphate capping enzyme |
| chr3_-_96634880 | 0.28 |
ENSMUST00000029741.9
|
Polr3c
|
polymerase (RNA) III (DNA directed) polypeptide C |
| chr5_+_108416763 | 0.27 |
ENSMUST00000031190.5
|
Dr1
|
down-regulator of transcription 1 |
| chr19_+_41818409 | 0.26 |
ENSMUST00000087155.5
|
Frat1
|
frequently rearranged in advanced T cell lymphomas |
| chr9_-_108067552 | 0.25 |
ENSMUST00000035208.14
|
Bsn
|
bassoon |
| chr16_-_22084700 | 0.23 |
ENSMUST00000161286.8
|
Tra2b
|
transformer 2 beta |
| chr7_+_44667377 | 0.22 |
ENSMUST00000044111.10
|
Rras
|
related RAS viral (r-ras) oncogene |
| chr1_-_75186067 | 0.22 |
ENSMUST00000186173.7
|
Glb1l
|
galactosidase, beta 1-like |
| chrX_+_134739783 | 0.20 |
ENSMUST00000173804.8
ENSMUST00000113136.8 |
Gprasp2
|
G protein-coupled receptor associated sorting protein 2 |
| chr7_+_79939747 | 0.20 |
ENSMUST00000205864.2
|
Vps33b
|
vacuolar protein sorting 33B |
| chr6_+_120643323 | 0.19 |
ENSMUST00000112686.8
|
Cecr2
|
CECR2, histone acetyl-lysine reader |
| chr9_-_44646487 | 0.18 |
ENSMUST00000034611.15
|
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr15_-_101759212 | 0.17 |
ENSMUST00000023790.5
|
Krt1
|
keratin 1 |
| chr3_-_135313982 | 0.16 |
ENSMUST00000132668.8
|
Nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 |
| chr7_-_19449319 | 0.15 |
ENSMUST00000032555.10
ENSMUST00000093552.12 |
Tomm40
|
translocase of outer mitochondrial membrane 40 |
| chr5_+_136937035 | 0.12 |
ENSMUST00000199101.5
ENSMUST00000200153.5 |
Ift22
|
intraflagellar transport 22 |
| chr2_-_84865831 | 0.12 |
ENSMUST00000028465.14
|
P2rx3
|
purinergic receptor P2X, ligand-gated ion channel, 3 |
| chr8_+_26401698 | 0.10 |
ENSMUST00000120653.8
ENSMUST00000126226.2 |
Kcnu1
|
potassium channel, subfamily U, member 1 |
| chr11_+_101066867 | 0.09 |
ENSMUST00000103109.4
|
Cntnap1
|
contactin associated protein-like 1 |
| chr11_+_75422925 | 0.09 |
ENSMUST00000169547.9
|
Slc43a2
|
solute carrier family 43, member 2 |
| chr5_+_136937083 | 0.09 |
ENSMUST00000008131.10
|
Ift22
|
intraflagellar transport 22 |
| chr12_+_102094977 | 0.09 |
ENSMUST00000159329.8
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr11_+_68322945 | 0.08 |
ENSMUST00000021283.8
|
Pik3r5
|
phosphoinositide-3-kinase regulatory subunit 5 |
| chr10_-_80223475 | 0.08 |
ENSMUST00000105350.3
|
Mex3d
|
mex3 RNA binding family member D |
| chr15_-_101588714 | 0.07 |
ENSMUST00000023786.7
|
Krt6b
|
keratin 6B |
| chr11_-_70578905 | 0.07 |
ENSMUST00000108544.8
|
Camta2
|
calmodulin binding transcription activator 2 |
| chr11_-_70895213 | 0.07 |
ENSMUST00000124464.2
ENSMUST00000108527.8 |
Dhx33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr7_+_135139542 | 0.07 |
ENSMUST00000073961.8
|
Ptpre
|
protein tyrosine phosphatase, receptor type, E |
| chr14_-_54855446 | 0.07 |
ENSMUST00000227257.2
ENSMUST00000022803.6 |
Psmb5
|
proteasome (prosome, macropain) subunit, beta type 5 |
| chr7_+_90125890 | 0.06 |
ENSMUST00000208919.2
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr7_+_79992839 | 0.05 |
ENSMUST00000032747.7
ENSMUST00000206480.2 ENSMUST00000206074.2 ENSMUST00000206122.2 |
Hddc3
|
HD domain containing 3 |
| chr19_+_8595369 | 0.03 |
ENSMUST00000010250.4
|
Slc22a6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr9_+_67747668 | 0.03 |
ENSMUST00000077879.7
|
Vps13c
|
vacuolar protein sorting 13C |
| chr11_-_70578744 | 0.03 |
ENSMUST00000108545.9
ENSMUST00000120261.8 |
Camta2
|
calmodulin binding transcription activator 2 |
| chr11_-_70578775 | 0.02 |
ENSMUST00000036299.14
ENSMUST00000119120.2 ENSMUST00000100933.10 |
Camta2
|
calmodulin binding transcription activator 2 |
| chr13_+_93441307 | 0.01 |
ENSMUST00000080127.12
|
Homer1
|
homer scaffolding protein 1 |
| chr3_+_133942244 | 0.01 |
ENSMUST00000181904.3
|
Cxxc4
|
CXXC finger 4 |
| chr14_+_54923655 | 0.01 |
ENSMUST00000038539.8
ENSMUST00000228027.2 |
1700123O20Rik
|
RIKEN cDNA 1700123O20 gene |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.4 | 1.4 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.3 | 1.2 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.3 | 0.8 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.2 | 2.7 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.2 | 0.6 | GO:0014737 | positive regulation of muscle atrophy(GO:0014737) |
| 0.2 | 2.9 | GO:0035646 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.2 | 1.3 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.2 | 1.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 0.3 | GO:0014728 | regulation of the force of skeletal muscle contraction(GO:0014728) regulation of skeletal muscle contraction by chemo-mechanical energy conversion(GO:0014862) |
| 0.1 | 5.2 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.1 | 3.1 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.1 | 0.8 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.1 | 0.6 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.9 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.1 | 0.2 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.1 | 0.7 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.1 | 0.4 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.1 | 0.5 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 0.2 | GO:1904629 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) response to diterpene(GO:1904629) cellular response to diterpene(GO:1904630) response to glucoside(GO:1904631) cellular response to glucoside(GO:1904632) |
| 0.0 | 2.2 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.7 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.8 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 3.7 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 1.5 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.1 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.0 | 0.7 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.2 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 1.6 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 1.1 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.2 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.6 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.2 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 3.0 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.5 | GO:0042092 | type 2 immune response(GO:0042092) |
| 0.0 | 1.3 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.4 | 1.2 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.4 | 2.2 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.2 | 1.6 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 1.6 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 2.7 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 0.2 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 0.1 | 0.5 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.1 | 0.2 | GO:0090537 | CERF complex(GO:0090537) |
| 0.1 | 1.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 2.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 2.7 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.4 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 1.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.4 | 1.4 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.2 | 1.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.2 | 1.6 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 0.9 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.1 | 1.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 5.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 1.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.6 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 1.1 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 0.5 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.4 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 0.8 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 6.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 3.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.2 | GO:0004527 | exonuclease activity(GO:0004527) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 3.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 2.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 2.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 2.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 3.3 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 5.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.1 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 1.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 0.9 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.6 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.6 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 1.7 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 1.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.3 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |