Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
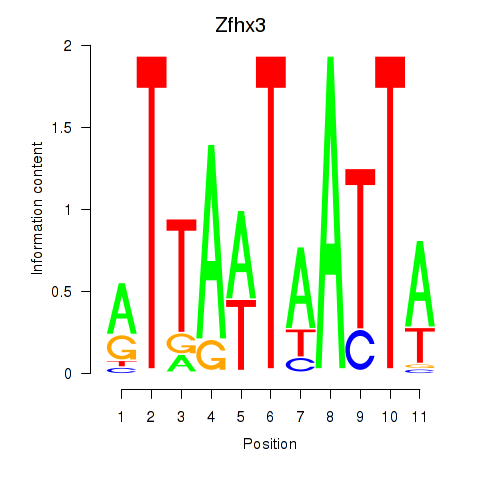
Results for Zfhx3
Z-value: 0.69
Transcription factors associated with Zfhx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfhx3
|
ENSMUSG00000038872.11 | Zfhx3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfhx3 | mm39_v1_chr8_+_109429732_109429732 | -0.17 | 3.3e-01 | Click! |
Activity profile of Zfhx3 motif
Sorted Z-values of Zfhx3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfhx3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_132318039 | 1.76 |
ENSMUST00000132435.2
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr5_+_90638580 | 1.74 |
ENSMUST00000042755.7
ENSMUST00000200693.2 |
Afp
|
alpha fetoprotein |
| chr1_+_174000304 | 1.55 |
ENSMUST00000027817.8
|
Spta1
|
spectrin alpha, erythrocytic 1 |
| chr17_-_31348576 | 1.52 |
ENSMUST00000024827.5
|
Tff3
|
trefoil factor 3, intestinal |
| chr15_-_60793115 | 1.46 |
ENSMUST00000096418.5
|
A1bg
|
alpha-1-B glycoprotein |
| chr19_+_9992380 | 1.41 |
ENSMUST00000137637.8
ENSMUST00000149967.9 |
Rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr10_+_79650496 | 1.09 |
ENSMUST00000218857.2
ENSMUST00000220365.2 |
Palm
|
paralemmin |
| chr9_+_96140781 | 1.06 |
ENSMUST00000190104.7
ENSMUST00000179416.8 ENSMUST00000189606.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr14_-_70945434 | 1.06 |
ENSMUST00000228346.2
|
Xpo7
|
exportin 7 |
| chr14_+_26722319 | 0.94 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr1_-_172722589 | 0.79 |
ENSMUST00000027824.7
|
Apcs
|
serum amyloid P-component |
| chr15_+_79578141 | 0.66 |
ENSMUST00000230898.2
ENSMUST00000229046.2 |
Gtpbp1
|
GTP binding protein 1 |
| chr11_+_24028022 | 0.64 |
ENSMUST00000000881.13
|
Bcl11a
|
B cell CLL/lymphoma 11A (zinc finger protein) |
| chr15_+_79575046 | 0.61 |
ENSMUST00000046463.10
|
Gtpbp1
|
GTP binding protein 1 |
| chr9_+_96140750 | 0.58 |
ENSMUST00000186609.7
|
Tfdp2
|
transcription factor Dp 2 |
| chr19_-_46661321 | 0.58 |
ENSMUST00000026012.8
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr11_+_24028111 | 0.54 |
ENSMUST00000109516.8
|
Bcl11a
|
B cell CLL/lymphoma 11A (zinc finger protein) |
| chr15_+_102367463 | 0.53 |
ENSMUST00000164938.8
ENSMUST00000023810.12 ENSMUST00000164957.8 ENSMUST00000171245.8 |
Prr13
|
proline rich 13 |
| chr19_-_46661501 | 0.52 |
ENSMUST00000236174.2
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr2_+_121337226 | 0.52 |
ENSMUST00000099473.10
ENSMUST00000110602.9 |
Wdr76
|
WD repeat domain 76 |
| chrX_+_162694397 | 0.51 |
ENSMUST00000140845.2
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr10_+_79762858 | 0.46 |
ENSMUST00000019708.12
ENSMUST00000105377.8 |
Arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr11_+_24028173 | 0.45 |
ENSMUST00000109514.8
|
Bcl11a
|
B cell CLL/lymphoma 11A (zinc finger protein) |
| chr3_-_103553347 | 0.43 |
ENSMUST00000117271.3
|
Atg4a-ps
|
autophagy related 4A, pseudogene |
| chr19_-_38032006 | 0.42 |
ENSMUST00000172095.3
ENSMUST00000041475.16 |
Myof
|
myoferlin |
| chr3_-_130524024 | 0.41 |
ENSMUST00000079085.11
|
Rpl34
|
ribosomal protein L34 |
| chr7_+_89814713 | 0.41 |
ENSMUST00000207084.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr5_-_44139099 | 0.39 |
ENSMUST00000061299.9
|
Fgfbp1
|
fibroblast growth factor binding protein 1 |
| chr18_+_11766333 | 0.39 |
ENSMUST00000115861.9
|
Rbbp8
|
retinoblastoma binding protein 8, endonuclease |
| chrX_+_162923474 | 0.33 |
ENSMUST00000073973.11
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr11_+_3933699 | 0.32 |
ENSMUST00000063004.14
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr5_-_44139121 | 0.32 |
ENSMUST00000199894.2
|
Fgfbp1
|
fibroblast growth factor binding protein 1 |
| chr19_-_38031774 | 0.31 |
ENSMUST00000226068.2
|
Myof
|
myoferlin |
| chr3_-_72875187 | 0.31 |
ENSMUST00000167334.8
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr14_+_75368532 | 0.31 |
ENSMUST00000143539.8
ENSMUST00000134114.8 |
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr9_-_119897328 | 0.30 |
ENSMUST00000177637.2
|
Cx3cr1
|
chemokine (C-X3-C motif) receptor 1 |
| chr14_+_75368939 | 0.29 |
ENSMUST00000125833.8
ENSMUST00000124499.8 |
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr3_+_84859453 | 0.28 |
ENSMUST00000029727.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr11_+_3933636 | 0.28 |
ENSMUST00000078757.8
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chrX_-_161747552 | 0.28 |
ENSMUST00000038769.3
|
S100g
|
S100 calcium binding protein G |
| chr1_-_126758369 | 0.27 |
ENSMUST00000112583.8
ENSMUST00000094609.10 |
Nckap5
|
NCK-associated protein 5 |
| chr9_-_119897358 | 0.27 |
ENSMUST00000064165.5
|
Cx3cr1
|
chemokine (C-X3-C motif) receptor 1 |
| chr3_+_153549846 | 0.27 |
ENSMUST00000044089.4
|
Asb17
|
ankyrin repeat and SOCS box-containing 17 |
| chr11_-_79421397 | 0.27 |
ENSMUST00000103236.4
ENSMUST00000170799.8 ENSMUST00000170422.4 |
Evi2a
Evi2
|
ecotropic viral integration site 2a ecotropic viral integration site 2 |
| chr14_-_86986541 | 0.27 |
ENSMUST00000226254.2
|
Diaph3
|
diaphanous related formin 3 |
| chr7_+_30193047 | 0.26 |
ENSMUST00000058280.13
ENSMUST00000133318.8 ENSMUST00000142575.8 ENSMUST00000131040.2 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chr9_+_111011327 | 0.25 |
ENSMUST00000216430.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr10_+_79763164 | 0.25 |
ENSMUST00000105376.2
|
Arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr6_-_141719536 | 0.25 |
ENSMUST00000148411.2
|
Gm5724
|
predicted gene 5724 |
| chr9_+_111011388 | 0.24 |
ENSMUST00000217117.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr15_-_98507913 | 0.24 |
ENSMUST00000226500.2
ENSMUST00000227501.2 |
Adcy6
|
adenylate cyclase 6 |
| chr9_+_113641615 | 0.23 |
ENSMUST00000111838.10
ENSMUST00000166734.10 ENSMUST00000214522.2 ENSMUST00000163895.3 |
Clasp2
|
CLIP associating protein 2 |
| chr6_+_142244145 | 0.23 |
ENSMUST00000041993.3
|
Iapp
|
islet amyloid polypeptide |
| chr4_+_109200225 | 0.23 |
ENSMUST00000030281.12
|
Eps15
|
epidermal growth factor receptor pathway substrate 15 |
| chr2_-_114005752 | 0.21 |
ENSMUST00000102543.5
ENSMUST00000043160.13 |
Aqr
|
aquarius |
| chr1_-_179572765 | 0.20 |
ENSMUST00000211943.3
ENSMUST00000131716.4 ENSMUST00000221136.2 |
Kif28
|
kinesin family member 28 |
| chr3_+_14545751 | 0.19 |
ENSMUST00000037321.8
ENSMUST00000120484.8 ENSMUST00000120801.2 |
Slc7a12
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr3_-_66204228 | 0.19 |
ENSMUST00000029419.8
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr1_+_125604706 | 0.19 |
ENSMUST00000027581.7
|
Gpr39
|
G protein-coupled receptor 39 |
| chr5_+_117378510 | 0.19 |
ENSMUST00000111975.3
|
Taok3
|
TAO kinase 3 |
| chr2_-_87868043 | 0.17 |
ENSMUST00000129056.3
|
Olfr73
|
olfactory receptor 73 |
| chr6_+_128376729 | 0.16 |
ENSMUST00000001561.12
|
Nrip2
|
nuclear receptor interacting protein 2 |
| chr13_+_109822527 | 0.16 |
ENSMUST00000074103.12
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr18_+_36661198 | 0.15 |
ENSMUST00000237174.2
ENSMUST00000236124.2 ENSMUST00000236779.2 ENSMUST00000235181.2 ENSMUST00000074298.13 ENSMUST00000115694.3 ENSMUST00000236126.2 |
Slc4a9
|
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
| chr1_-_158642039 | 0.15 |
ENSMUST00000161589.3
|
Pappa2
|
pappalysin 2 |
| chr3_+_82933383 | 0.14 |
ENSMUST00000029630.15
ENSMUST00000166581.4 |
Fga
|
fibrinogen alpha chain |
| chr13_-_76091931 | 0.13 |
ENSMUST00000022078.12
ENSMUST00000109606.3 |
Rhobtb3
|
Rho-related BTB domain containing 3 |
| chr15_-_55411560 | 0.13 |
ENSMUST00000165356.2
|
Mrpl13
|
mitochondrial ribosomal protein L13 |
| chr17_+_35960600 | 0.13 |
ENSMUST00000171166.3
|
Sfta2
|
surfactant associated 2 |
| chr6_-_30936013 | 0.13 |
ENSMUST00000101589.5
|
Klf14
|
Kruppel-like factor 14 |
| chr19_-_33764859 | 0.13 |
ENSMUST00000148137.9
|
Lipo1
|
lipase, member O1 |
| chr1_-_138103021 | 0.12 |
ENSMUST00000182755.8
ENSMUST00000193650.2 ENSMUST00000182283.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr8_-_66939146 | 0.12 |
ENSMUST00000026681.7
|
Tma16
|
translation machinery associated 16 |
| chr5_-_137784912 | 0.12 |
ENSMUST00000031740.16
|
Mepce
|
methylphosphate capping enzyme |
| chr18_-_66155651 | 0.12 |
ENSMUST00000143990.2
|
Lman1
|
lectin, mannose-binding, 1 |
| chr4_+_101365144 | 0.11 |
ENSMUST00000149047.8
ENSMUST00000106929.10 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr10_-_88440869 | 0.10 |
ENSMUST00000119185.8
ENSMUST00000238199.2 |
Mybpc1
|
myosin binding protein C, slow-type |
| chr7_-_100232276 | 0.10 |
ENSMUST00000152876.3
ENSMUST00000150042.8 ENSMUST00000132888.9 |
Mrpl48
|
mitochondrial ribosomal protein L48 |
| chr3_+_75982890 | 0.09 |
ENSMUST00000160261.8
|
Fstl5
|
follistatin-like 5 |
| chr2_+_89821565 | 0.09 |
ENSMUST00000111509.3
ENSMUST00000213909.3 |
Olfr1261
|
olfactory receptor 1261 |
| chr15_+_39255185 | 0.09 |
ENSMUST00000228839.2
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr11_-_41891111 | 0.09 |
ENSMUST00000109290.2
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
| chr8_+_94537910 | 0.09 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr4_+_109092610 | 0.09 |
ENSMUST00000106628.8
|
Calr4
|
calreticulin 4 |
| chr1_-_138102972 | 0.08 |
ENSMUST00000195533.6
ENSMUST00000183301.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr14_-_50479161 | 0.08 |
ENSMUST00000214388.2
|
Olfr731
|
olfactory receptor 731 |
| chr7_+_103578032 | 0.08 |
ENSMUST00000106863.2
|
Olfr631
|
olfactory receptor 631 |
| chr2_+_109522781 | 0.08 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr14_+_8283087 | 0.08 |
ENSMUST00000206298.3
ENSMUST00000216079.2 |
Olfr720
|
olfactory receptor 720 |
| chr3_-_144638284 | 0.08 |
ENSMUST00000098549.4
|
Clca4b
|
chloride channel accessory 4B |
| chr6_+_128376844 | 0.08 |
ENSMUST00000120405.4
|
Nrip2
|
nuclear receptor interacting protein 2 |
| chr4_+_109092459 | 0.08 |
ENSMUST00000106631.9
|
Calr4
|
calreticulin 4 |
| chr12_-_8589545 | 0.07 |
ENSMUST00000095863.10
ENSMUST00000165657.3 |
Slc7a15
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 15 |
| chr3_+_60380463 | 0.07 |
ENSMUST00000195077.6
ENSMUST00000193647.6 ENSMUST00000195001.2 ENSMUST00000192807.6 |
Mbnl1
|
muscleblind like splicing factor 1 |
| chr11_+_54517164 | 0.07 |
ENSMUST00000239168.2
|
Rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr10_-_130230201 | 0.07 |
ENSMUST00000094502.6
ENSMUST00000233146.2 |
Vmn2r84
|
vomeronasal 2, receptor 84 |
| chr2_+_110427643 | 0.06 |
ENSMUST00000045972.13
ENSMUST00000111026.3 |
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12 |
| chr6_-_122317457 | 0.06 |
ENSMUST00000160843.8
|
Phc1
|
polyhomeotic 1 |
| chr14_-_50479323 | 0.06 |
ENSMUST00000214152.2
|
Olfr731
|
olfactory receptor 731 |
| chr2_-_87467879 | 0.06 |
ENSMUST00000216082.2
|
Olfr1132
|
olfactory receptor 1132 |
| chr1_-_66984521 | 0.06 |
ENSMUST00000160100.2
|
Myl1
|
myosin, light polypeptide 1 |
| chr1_-_135934080 | 0.06 |
ENSMUST00000166193.9
|
Igfn1
|
immunoglobulin-like and fibronectin type III domain containing 1 |
| chr2_+_85648823 | 0.06 |
ENSMUST00000214416.2
|
Olfr1018
|
olfactory receptor 1018 |
| chr3_+_7494108 | 0.06 |
ENSMUST00000193330.2
|
Pkia
|
protein kinase inhibitor, alpha |
| chrM_+_2743 | 0.06 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr17_-_43813664 | 0.05 |
ENSMUST00000024707.9
ENSMUST00000117137.8 |
Mep1a
|
meprin 1 alpha |
| chr5_+_38233901 | 0.05 |
ENSMUST00000146864.6
|
Stx18
|
syntaxin 18 |
| chr5_-_123804745 | 0.05 |
ENSMUST00000149410.2
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr1_-_126758520 | 0.05 |
ENSMUST00000162646.8
|
Nckap5
|
NCK-associated protein 5 |
| chr3_-_59127571 | 0.05 |
ENSMUST00000199675.2
ENSMUST00000170388.6 |
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr10_+_129184719 | 0.05 |
ENSMUST00000213970.2
|
Olfr782
|
olfactory receptor 782 |
| chr7_-_28661751 | 0.05 |
ENSMUST00000068045.14
ENSMUST00000217157.2 |
Actn4
|
actinin alpha 4 |
| chr7_-_101486983 | 0.05 |
ENSMUST00000185929.2
ENSMUST00000165052.8 |
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr2_-_140513320 | 0.04 |
ENSMUST00000056760.4
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr9_+_53678801 | 0.04 |
ENSMUST00000048670.10
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr6_+_65502344 | 0.04 |
ENSMUST00000212375.2
|
Tnip3
|
TNFAIP3 interacting protein 3 |
| chr4_-_154721288 | 0.04 |
ENSMUST00000030902.13
ENSMUST00000105637.8 ENSMUST00000070313.14 ENSMUST00000105636.8 ENSMUST00000105638.9 ENSMUST00000097759.9 ENSMUST00000124771.2 |
Prdm16
|
PR domain containing 16 |
| chr2_-_140513382 | 0.04 |
ENSMUST00000110057.3
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr6_-_23650205 | 0.04 |
ENSMUST00000115354.2
|
Rnf133
|
ring finger protein 133 |
| chr3_-_98364359 | 0.04 |
ENSMUST00000188356.3
ENSMUST00000167753.8 |
Gm4450
|
predicted gene 4450 |
| chr18_-_84854841 | 0.04 |
ENSMUST00000236689.2
|
Gm17266
|
predicted gene, 17266 |
| chrX_+_111221031 | 0.04 |
ENSMUST00000026599.10
ENSMUST00000113415.2 |
Apool
|
apolipoprotein O-like |
| chr14_+_8282925 | 0.04 |
ENSMUST00000217642.2
|
Olfr720
|
olfactory receptor 720 |
| chr7_-_66915756 | 0.04 |
ENSMUST00000207715.2
|
Mef2a
|
myocyte enhancer factor 2A |
| chr18_+_37102969 | 0.04 |
ENSMUST00000194544.2
|
Gm37013
|
predicted gene, 37013 |
| chr2_+_86122799 | 0.03 |
ENSMUST00000217166.2
|
Olfr1052
|
olfactory receptor 1052 |
| chr8_+_69661683 | 0.03 |
ENSMUST00000212681.2
ENSMUST00000212312.2 |
Zfp930
|
zinc finger protein 930 |
| chr3_-_69767208 | 0.03 |
ENSMUST00000171529.4
ENSMUST00000051239.13 |
Sptssb
|
serine palmitoyltransferase, small subunit B |
| chr1_+_179788675 | 0.03 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr10_+_23672842 | 0.03 |
ENSMUST00000119597.8
ENSMUST00000179321.8 ENSMUST00000133289.2 |
Slc18b1
|
solute carrier family 18, subfamily B, member 1 |
| chr2_-_111880531 | 0.03 |
ENSMUST00000213582.2
ENSMUST00000213961.3 ENSMUST00000215531.2 |
Olfr1312
|
olfactory receptor 1312 |
| chr3_-_144514386 | 0.03 |
ENSMUST00000197013.2
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr6_-_86742789 | 0.03 |
ENSMUST00000123732.4
|
Anxa4
|
annexin A4 |
| chr1_+_172525613 | 0.03 |
ENSMUST00000038495.5
|
Crp
|
C-reactive protein, pentraxin-related |
| chr12_+_86999366 | 0.03 |
ENSMUST00000191463.2
|
Cipc
|
CLOCK interacting protein, circadian |
| chr5_-_87638728 | 0.03 |
ENSMUST00000147854.6
|
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr7_+_23274951 | 0.03 |
ENSMUST00000228832.2
ENSMUST00000227547.2 ENSMUST00000226669.2 ENSMUST00000227932.2 |
Vmn1r169
|
vomeronasal 1 receptor 169 |
| chr9_-_48876290 | 0.03 |
ENSMUST00000008734.5
|
Htr3b
|
5-hydroxytryptamine (serotonin) receptor 3B |
| chr7_-_144305705 | 0.02 |
ENSMUST00000155175.8
|
Ano1
|
anoctamin 1, calcium activated chloride channel |
| chr7_-_10011933 | 0.02 |
ENSMUST00000227719.2
ENSMUST00000228622.2 ENSMUST00000228086.2 |
Vmn1r66
|
vomeronasal 1 receptor 66 |
| chr5_-_137784943 | 0.02 |
ENSMUST00000132726.2
|
Mepce
|
methylphosphate capping enzyme |
| chr3_+_125197722 | 0.02 |
ENSMUST00000173932.8
|
Ndst4
|
N-deacetylase/N-sulfotransferase (heparin glucosaminyl) 4 |
| chr1_+_179788037 | 0.02 |
ENSMUST00000097453.9
ENSMUST00000111117.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr17_+_38143840 | 0.02 |
ENSMUST00000213857.2
|
Olfr125
|
olfactory receptor 125 |
| chr2_+_67004178 | 0.02 |
ENSMUST00000239009.2
ENSMUST00000238912.2 |
Xirp2
|
xin actin-binding repeat containing 2 |
| chr2_-_37537224 | 0.02 |
ENSMUST00000028279.10
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr3_+_66892979 | 0.02 |
ENSMUST00000162362.8
ENSMUST00000065074.14 ENSMUST00000065047.13 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chrM_+_7006 | 0.02 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr18_+_37651393 | 0.02 |
ENSMUST00000097609.3
|
Pcdhb22
|
protocadherin beta 22 |
| chr12_-_101785307 | 0.02 |
ENSMUST00000021603.9
|
Fbln5
|
fibulin 5 |
| chr2_-_89195205 | 0.02 |
ENSMUST00000111543.2
ENSMUST00000137692.3 |
Olfr1234
|
olfactory receptor 1234 |
| chr16_-_56748424 | 0.02 |
ENSMUST00000210579.2
|
Lnp1
|
leukemia NUP98 fusion partner 1 |
| chr12_+_84463970 | 0.01 |
ENSMUST00000183146.2
|
Rnf113a2
|
ring finger protein 113A2 |
| chr7_-_107633196 | 0.01 |
ENSMUST00000210173.3
|
Olfr478
|
olfactory receptor 478 |
| chrX_-_142716200 | 0.01 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr9_+_39903409 | 0.01 |
ENSMUST00000217600.2
|
Olfr978
|
olfactory receptor 978 |
| chr1_-_4430481 | 0.00 |
ENSMUST00000027032.6
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr2_-_88581690 | 0.00 |
ENSMUST00000215179.3
ENSMUST00000215529.3 |
Olfr1198
|
olfactory receptor 1198 |
| chr7_-_23536797 | 0.00 |
ENSMUST00000226319.2
ENSMUST00000228793.2 ENSMUST00000228280.2 ENSMUST00000227661.2 ENSMUST00000226767.2 ENSMUST00000227129.2 |
Vmn1r176
|
vomeronasal 1 receptor 176 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:1904800 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) |
| 0.3 | 0.8 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.2 | 0.6 | GO:0002880 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.2 | 1.1 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.1 | 2.8 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.1 | 0.5 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.1 | 0.9 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.1 | 0.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.3 | GO:0015827 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.1 | 0.2 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.1 | 0.7 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 0.4 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 0.6 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.3 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 0.2 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.0 | 0.4 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.7 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0035483 | gastric emptying(GO:0035483) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 1.6 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 1.3 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.2 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.1 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.6 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 1.8 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.5 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.2 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.1 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.1 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.1 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 0.0 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.0 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.0 | 1.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.1 | GO:0044861 | protein localization to plasma membrane raft(GO:0044860) protein transport into plasma membrane raft(GO:0044861) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 1.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 1.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.2 | GO:1903754 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.1 | 0.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 0.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.2 | 1.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.1 | 0.6 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.1 | 0.8 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 1.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.3 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 0.6 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 0.3 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 1.1 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.3 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.4 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 1.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.7 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.3 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 1.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 1.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |