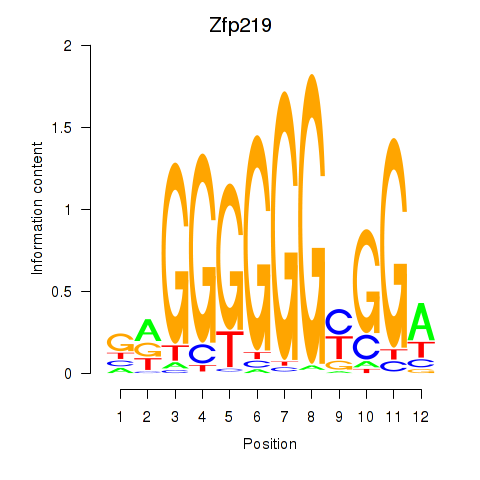
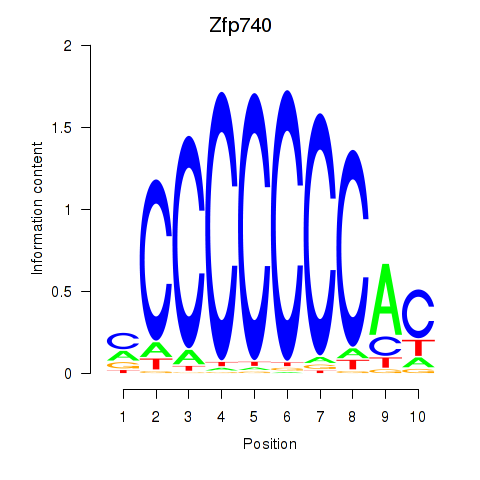
Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Zfp219_Zfp740
Z-value: 1.20
Transcription factors associated with Zfp219_Zfp740
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp219
|
ENSMUSG00000049295.18 | Zfp219 |
|
Zfp740
|
ENSMUSG00000046897.18 | Zfp740 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp219 | mm39_v1_chr14_-_52252318_52252369 | -0.29 | 8.4e-02 | Click! |
| Zfp740 | mm39_v1_chr15_+_102112074_102112144 | 0.17 | 3.2e-01 | Click! |
Activity profile of Zfp219_Zfp740 motif
Sorted Z-values of Zfp219_Zfp740 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp219_Zfp740
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_129326004 | 2.21 |
ENSMUST00000199910.5
ENSMUST00000197070.5 ENSMUST00000071402.7 |
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr3_+_95836558 | 2.01 |
ENSMUST00000165307.8
ENSMUST00000015893.13 ENSMUST00000169426.8 |
Anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr3_+_95836637 | 1.88 |
ENSMUST00000171368.8
ENSMUST00000168106.8 |
Anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr12_+_109419371 | 1.76 |
ENSMUST00000109844.11
ENSMUST00000109842.9 ENSMUST00000109843.8 |
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr7_-_126626152 | 1.72 |
ENSMUST00000206254.2
ENSMUST00000206291.2 |
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr4_-_87951565 | 1.69 |
ENSMUST00000078090.12
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr12_+_109419575 | 1.63 |
ENSMUST00000173539.8
ENSMUST00000109841.9 |
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr5_+_118698689 | 1.49 |
ENSMUST00000100816.8
ENSMUST00000201010.2 |
Med13l
|
mediator complex subunit 13-like |
| chr7_-_126625739 | 1.44 |
ENSMUST00000205461.2
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr12_+_109419454 | 1.41 |
ENSMUST00000109846.11
|
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr7_-_126625657 | 1.39 |
ENSMUST00000205568.2
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr7_-_126625617 | 1.39 |
ENSMUST00000032916.6
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr11_+_61967821 | 1.38 |
ENSMUST00000092415.9
ENSMUST00000201015.4 ENSMUST00000202744.4 ENSMUST00000201723.4 ENSMUST00000202179.2 |
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chrX_+_100298134 | 1.36 |
ENSMUST00000062000.6
|
Foxo4
|
forkhead box O4 |
| chr2_+_103800553 | 1.32 |
ENSMUST00000111140.3
ENSMUST00000111139.3 |
Lmo2
|
LIM domain only 2 |
| chr7_+_89779564 | 1.31 |
ENSMUST00000208742.2
ENSMUST00000049537.9 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr11_-_88609048 | 1.30 |
ENSMUST00000107909.8
|
Msi2
|
musashi RNA-binding protein 2 |
| chr7_+_89779421 | 1.19 |
ENSMUST00000207225.2
ENSMUST00000207484.2 ENSMUST00000209068.2 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chrX_-_48877080 | 1.18 |
ENSMUST00000114893.8
|
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr14_-_70864448 | 1.15 |
ENSMUST00000110984.4
|
Dmtn
|
dematin actin binding protein |
| chr7_+_127376550 | 1.11 |
ENSMUST00000126761.8
ENSMUST00000047157.13 |
Setd1a
|
SET domain containing 1A |
| chr10_+_43455157 | 1.10 |
ENSMUST00000058714.10
|
Cd24a
|
CD24a antigen |
| chr14_-_70864666 | 1.10 |
ENSMUST00000022694.17
|
Dmtn
|
dematin actin binding protein |
| chr4_+_129355374 | 1.08 |
ENSMUST00000048162.10
ENSMUST00000138013.3 |
Bsdc1
|
BSD domain containing 1 |
| chr11_+_94881861 | 1.06 |
ENSMUST00000038696.12
|
Ppp1r9b
|
protein phosphatase 1, regulatory subunit 9B |
| chr2_+_84564394 | 1.03 |
ENSMUST00000238573.2
ENSMUST00000090729.9 |
Ypel4
|
yippee like 4 |
| chr7_+_89779493 | 1.02 |
ENSMUST00000208730.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr11_+_106916430 | 1.01 |
ENSMUST00000140447.4
|
1810010H24Rik
|
RIKEN cDNA 1810010H24 gene |
| chr12_-_118265103 | 0.99 |
ENSMUST00000222314.2
ENSMUST00000026367.11 |
Sp4
|
trans-acting transcription factor 4 |
| chr11_-_88608920 | 0.98 |
ENSMUST00000092794.12
|
Msi2
|
musashi RNA-binding protein 2 |
| chr14_-_71003973 | 0.97 |
ENSMUST00000226448.2
ENSMUST00000022696.8 |
Xpo7
|
exportin 7 |
| chrX_-_101294545 | 0.96 |
ENSMUST00000134887.8
|
Cited1
|
Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 |
| chr4_-_133746138 | 0.96 |
ENSMUST00000051674.3
|
Lin28a
|
lin-28 homolog A (C. elegans) |
| chr6_-_86646118 | 0.95 |
ENSMUST00000001184.10
|
Mxd1
|
MAX dimerization protein 1 |
| chr2_+_103800459 | 0.94 |
ENSMUST00000111143.8
ENSMUST00000138815.2 |
Lmo2
|
LIM domain only 2 |
| chr10_+_11157326 | 0.94 |
ENSMUST00000070300.5
|
Fbxo30
|
F-box protein 30 |
| chr12_-_4891435 | 0.92 |
ENSMUST00000219880.2
ENSMUST00000020964.7 |
Fkbp1b
|
FK506 binding protein 1b |
| chr7_+_127376267 | 0.92 |
ENSMUST00000144406.8
|
Setd1a
|
SET domain containing 1A |
| chr3_+_129326285 | 0.91 |
ENSMUST00000197235.5
|
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr10_+_11157047 | 0.88 |
ENSMUST00000129456.8
|
Fbxo30
|
F-box protein 30 |
| chr11_-_69872050 | 0.87 |
ENSMUST00000108594.8
|
Elp5
|
elongator acetyltransferase complex subunit 5 |
| chr7_-_43182595 | 0.87 |
ENSMUST00000205503.2
|
Cd33
|
CD33 antigen |
| chr14_-_71004019 | 0.86 |
ENSMUST00000167242.8
|
Xpo7
|
exportin 7 |
| chr14_-_70866385 | 0.86 |
ENSMUST00000228824.2
|
Dmtn
|
dematin actin binding protein |
| chr15_+_81469538 | 0.84 |
ENSMUST00000068387.11
|
Ep300
|
E1A binding protein p300 |
| chr11_+_117740077 | 0.81 |
ENSMUST00000081387.11
|
Birc5
|
baculoviral IAP repeat-containing 5 |
| chr11_-_102208615 | 0.80 |
ENSMUST00000107117.9
|
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr15_-_89310060 | 0.78 |
ENSMUST00000109313.9
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chrX_-_37653396 | 0.78 |
ENSMUST00000016681.15
|
Cul4b
|
cullin 4B |
| chr8_+_85696695 | 0.76 |
ENSMUST00000164807.2
|
Prdx2
|
peroxiredoxin 2 |
| chrX_+_41238193 | 0.75 |
ENSMUST00000115073.9
ENSMUST00000115072.8 |
Stag2
|
stromal antigen 2 |
| chr11_+_23206001 | 0.75 |
ENSMUST00000020538.13
ENSMUST00000109551.8 ENSMUST00000102870.8 ENSMUST00000102869.8 |
Xpo1
|
exportin 1 |
| chr11_+_98094134 | 0.74 |
ENSMUST00000003203.14
ENSMUST00000107538.2 |
Cdk12
|
cyclin-dependent kinase 12 |
| chr5_+_32616187 | 0.74 |
ENSMUST00000015100.15
|
Ppp1cb
|
protein phosphatase 1 catalytic subunit beta |
| chr10_+_20188207 | 0.72 |
ENSMUST00000092678.10
ENSMUST00000043881.12 |
Bclaf1
|
BCL2-associated transcription factor 1 |
| chr8_+_85696396 | 0.71 |
ENSMUST00000109733.8
|
Prdx2
|
peroxiredoxin 2 |
| chr8_+_85696453 | 0.71 |
ENSMUST00000125893.8
|
Prdx2
|
peroxiredoxin 2 |
| chr2_+_78699360 | 0.71 |
ENSMUST00000028398.14
|
Ube2e3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr3_+_87813624 | 0.69 |
ENSMUST00000005017.15
|
Hdgf
|
heparin binding growth factor |
| chr8_+_85696216 | 0.69 |
ENSMUST00000109734.8
ENSMUST00000005292.15 |
Prdx2
|
peroxiredoxin 2 |
| chr9_-_66826928 | 0.68 |
ENSMUST00000041139.9
|
Rab8b
|
RAB8B, member RAS oncogene family |
| chr4_+_41135730 | 0.68 |
ENSMUST00000040008.4
|
Ube2r2
|
ubiquitin-conjugating enzyme E2R 2 |
| chr13_+_91609169 | 0.65 |
ENSMUST00000004094.15
ENSMUST00000042122.15 |
Ssbp2
|
single-stranded DNA binding protein 2 |
| chr11_-_102208449 | 0.63 |
ENSMUST00000107123.10
|
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr3_+_28317570 | 0.62 |
ENSMUST00000160307.9
ENSMUST00000159680.9 ENSMUST00000160518.8 ENSMUST00000162485.8 ENSMUST00000159308.8 ENSMUST00000162777.8 ENSMUST00000161964.2 |
Tnik
|
TRAF2 and NCK interacting kinase |
| chr17_+_34811217 | 0.62 |
ENSMUST00000038149.13
|
Pbx2
|
pre B cell leukemia homeobox 2 |
| chr2_+_117942357 | 0.62 |
ENSMUST00000039559.9
|
Thbs1
|
thrombospondin 1 |
| chr11_+_3239868 | 0.60 |
ENSMUST00000094471.10
ENSMUST00000110043.8 |
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr18_+_34910064 | 0.60 |
ENSMUST00000043775.9
ENSMUST00000224715.2 |
Kdm3b
|
KDM3B lysine (K)-specific demethylase 3B |
| chr1_-_84817000 | 0.59 |
ENSMUST00000186648.7
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr15_-_76554065 | 0.59 |
ENSMUST00000037824.6
|
Foxh1
|
forkhead box H1 |
| chr7_-_126736979 | 0.58 |
ENSMUST00000049931.6
|
Spn
|
sialophorin |
| chr10_+_20188269 | 0.58 |
ENSMUST00000185800.7
|
Bclaf1
|
BCL2-associated transcription factor 1 |
| chr13_+_75855695 | 0.56 |
ENSMUST00000222194.2
ENSMUST00000223535.2 ENSMUST00000222853.2 |
Ell2
|
elongation factor for RNA polymerase II 2 |
| chr11_+_72851989 | 0.56 |
ENSMUST00000163326.8
ENSMUST00000108485.9 ENSMUST00000021142.8 ENSMUST00000108486.8 ENSMUST00000108484.8 |
Atp2a3
|
ATPase, Ca++ transporting, ubiquitous |
| chr5_+_32616566 | 0.55 |
ENSMUST00000202078.2
|
Ppp1cb
|
protein phosphatase 1 catalytic subunit beta |
| chr4_-_59549243 | 0.55 |
ENSMUST00000173699.8
ENSMUST00000173884.8 ENSMUST00000102883.11 ENSMUST00000174586.8 |
Ptbp3
|
polypyrimidine tract binding protein 3 |
| chrX_-_101294380 | 0.55 |
ENSMUST00000130589.2
|
Cited1
|
Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 |
| chr11_-_85030761 | 0.54 |
ENSMUST00000108075.9
|
Usp32
|
ubiquitin specific peptidase 32 |
| chr6_+_51447613 | 0.54 |
ENSMUST00000114445.8
ENSMUST00000114446.8 ENSMUST00000141711.3 |
Cbx3
|
chromobox 3 |
| chr15_+_96185399 | 0.54 |
ENSMUST00000134985.9
ENSMUST00000096250.5 |
Arid2
|
AT rich interactive domain 2 (ARID, RFX-like) |
| chr18_-_10030017 | 0.54 |
ENSMUST00000116669.2
ENSMUST00000092096.14 |
Usp14
|
ubiquitin specific peptidase 14 |
| chr5_+_97145533 | 0.54 |
ENSMUST00000112974.6
ENSMUST00000035635.10 |
Bmp2k
|
BMP2 inducible kinase |
| chr8_-_85413707 | 0.53 |
ENSMUST00000238301.2
|
Nacc1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr6_+_120643323 | 0.53 |
ENSMUST00000112686.8
|
Cecr2
|
CECR2, histone acetyl-lysine reader |
| chr7_+_90091937 | 0.53 |
ENSMUST00000061767.5
|
Crebzf
|
CREB/ATF bZIP transcription factor |
| chr10_+_79833296 | 0.53 |
ENSMUST00000171637.8
ENSMUST00000043866.8 |
Abca7
|
ATP-binding cassette, sub-family A (ABC1), member 7 |
| chr17_-_47169380 | 0.53 |
ENSMUST00000233455.2
|
Bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr17_+_47905553 | 0.52 |
ENSMUST00000182846.3
|
Ccnd3
|
cyclin D3 |
| chr2_+_75489596 | 0.52 |
ENSMUST00000111964.8
ENSMUST00000111962.8 ENSMUST00000111961.8 ENSMUST00000164947.9 ENSMUST00000090792.11 |
Hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr7_+_127345909 | 0.51 |
ENSMUST00000033081.14
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr5_-_123126550 | 0.50 |
ENSMUST00000086200.11
ENSMUST00000156474.8 |
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr5_+_122296322 | 0.50 |
ENSMUST00000102528.11
ENSMUST00000086294.11 |
Ppp1cc
|
protein phosphatase 1 catalytic subunit gamma |
| chr13_+_91609264 | 0.50 |
ENSMUST00000231481.2
|
Ssbp2
|
single-stranded DNA binding protein 2 |
| chr4_+_8691303 | 0.48 |
ENSMUST00000051558.10
|
Chd7
|
chromodomain helicase DNA binding protein 7 |
| chr2_+_174171799 | 0.47 |
ENSMUST00000109085.8
|
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chrX_+_41239548 | 0.47 |
ENSMUST00000069619.14
|
Stag2
|
stromal antigen 2 |
| chr5_-_24597009 | 0.46 |
ENSMUST00000059401.7
|
Atg9b
|
autophagy related 9B |
| chr6_+_67243967 | 0.46 |
ENSMUST00000203436.3
ENSMUST00000205106.3 ENSMUST00000204293.3 ENSMUST00000203077.3 ENSMUST00000204294.3 |
Serbp1
|
serpine1 mRNA binding protein 1 |
| chr3_+_60503051 | 0.46 |
ENSMUST00000192757.6
ENSMUST00000193518.6 ENSMUST00000195817.3 |
Mbnl1
|
muscleblind like splicing factor 1 |
| chr4_-_136563154 | 0.46 |
ENSMUST00000105846.9
ENSMUST00000059287.14 ENSMUST00000105845.9 |
Ephb2
|
Eph receptor B2 |
| chr11_+_77873535 | 0.45 |
ENSMUST00000108360.8
ENSMUST00000049167.14 |
Phf12
|
PHD finger protein 12 |
| chr19_-_46033353 | 0.45 |
ENSMUST00000026252.14
ENSMUST00000156585.9 ENSMUST00000185355.7 ENSMUST00000152946.8 |
Ldb1
|
LIM domain binding 1 |
| chr7_-_132415257 | 0.45 |
ENSMUST00000097999.9
|
Fam53b
|
family with sequence similarity 53, member B |
| chrX_+_70600481 | 0.45 |
ENSMUST00000123100.2
|
Hmgb3
|
high mobility group box 3 |
| chr1_-_84817022 | 0.44 |
ENSMUST00000189496.7
ENSMUST00000027421.13 ENSMUST00000186894.7 |
Trip12
|
thyroid hormone receptor interactor 12 |
| chr3_+_130904000 | 0.44 |
ENSMUST00000029611.14
ENSMUST00000106341.9 ENSMUST00000066849.13 |
Lef1
|
lymphoid enhancer binding factor 1 |
| chr1_-_180641159 | 0.44 |
ENSMUST00000162118.8
ENSMUST00000159685.2 ENSMUST00000161308.8 |
H3f3a
|
H3.3 histone A |
| chr19_-_45804446 | 0.44 |
ENSMUST00000079431.10
ENSMUST00000026247.13 ENSMUST00000162528.9 |
Kcnip2
|
Kv channel-interacting protein 2 |
| chr11_+_97340962 | 0.44 |
ENSMUST00000107601.8
|
Arhgap23
|
Rho GTPase activating protein 23 |
| chr3_+_138232176 | 0.44 |
ENSMUST00000200020.5
|
Eif4e
|
eukaryotic translation initiation factor 4E |
| chr14_+_54713703 | 0.43 |
ENSMUST00000164697.8
|
Rem2
|
rad and gem related GTP binding protein 2 |
| chr2_+_109111083 | 0.43 |
ENSMUST00000028527.8
|
Kif18a
|
kinesin family member 18A |
| chr15_-_103123711 | 0.42 |
ENSMUST00000122182.2
ENSMUST00000108813.10 ENSMUST00000127191.2 |
Cbx5
|
chromobox 5 |
| chr10_-_123032821 | 0.42 |
ENSMUST00000219619.2
ENSMUST00000020334.9 |
Usp15
|
ubiquitin specific peptidase 15 |
| chr6_+_124986627 | 0.42 |
ENSMUST00000046064.17
ENSMUST00000152752.8 ENSMUST00000088308.10 ENSMUST00000112425.8 ENSMUST00000084275.12 |
Zfp384
|
zinc finger protein 384 |
| chr1_-_180641099 | 0.41 |
ENSMUST00000159789.2
ENSMUST00000081026.11 |
H3f3a
|
H3.3 histone A |
| chr10_+_74802996 | 0.41 |
ENSMUST00000037813.5
|
Gnaz
|
guanine nucleotide binding protein, alpha z subunit |
| chr1_+_171097891 | 0.41 |
ENSMUST00000064272.10
ENSMUST00000151863.8 ENSMUST00000141999.8 ENSMUST00000111313.10 ENSMUST00000126699.4 |
B4galt3
|
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 3 |
| chr11_+_96173355 | 0.41 |
ENSMUST00000125410.2
|
Hoxb8
|
homeobox B8 |
| chr2_-_169973076 | 0.40 |
ENSMUST00000063710.13
|
Zfp217
|
zinc finger protein 217 |
| chr1_-_180641430 | 0.40 |
ENSMUST00000162814.8
|
H3f3a
|
H3.3 histone A |
| chr17_-_28039588 | 0.40 |
ENSMUST00000114863.10
ENSMUST00000233131.2 |
Ilrun
|
inflammation and lipid regulator with UBA-like and NBR1-like domains |
| chr2_+_174171860 | 0.40 |
ENSMUST00000109087.8
ENSMUST00000109084.8 |
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr2_-_153286361 | 0.39 |
ENSMUST00000109784.2
|
Nol4l
|
nucleolar protein 4-like |
| chr9_+_72439891 | 0.39 |
ENSMUST00000183372.8
ENSMUST00000184015.2 |
Rfx7
|
regulatory factor X, 7 |
| chr5_+_3853160 | 0.38 |
ENSMUST00000080085.9
ENSMUST00000200386.5 |
Krit1
|
KRIT1, ankyrin repeat containing |
| chr9_-_21982637 | 0.38 |
ENSMUST00000179605.9
ENSMUST00000043922.7 |
Zfp653
|
zinc finger protein 653 |
| chr4_+_32615428 | 0.38 |
ENSMUST00000178925.8
ENSMUST00000029950.10 |
Casp8ap2
|
caspase 8 associated protein 2 |
| chr3_+_96069271 | 0.38 |
ENSMUST00000054356.16
|
Mtmr11
|
myotubularin related protein 11 |
| chr2_-_121101822 | 0.38 |
ENSMUST00000110647.8
ENSMUST00000110648.8 |
Trp53bp1
|
transformation related protein 53 binding protein 1 |
| chr11_+_69871952 | 0.38 |
ENSMUST00000108593.8
|
Ctdnep1
|
CTD nuclear envelope phosphatase 1 |
| chr11_+_97306353 | 0.38 |
ENSMUST00000121799.8
|
Arhgap23
|
Rho GTPase activating protein 23 |
| chr11_-_88608958 | 0.37 |
ENSMUST00000107908.2
|
Msi2
|
musashi RNA-binding protein 2 |
| chr17_-_32607859 | 0.37 |
ENSMUST00000087703.12
ENSMUST00000170603.3 |
Wiz
|
widely-interspaced zinc finger motifs |
| chr6_-_28261881 | 0.37 |
ENSMUST00000115320.8
ENSMUST00000123098.8 ENSMUST00000115321.9 ENSMUST00000155494.2 |
Zfp800
|
zinc finger protein 800 |
| chr11_+_103061905 | 0.37 |
ENSMUST00000042286.12
ENSMUST00000218163.2 |
Fmnl1
|
formin-like 1 |
| chr17_-_28039506 | 0.37 |
ENSMUST00000114859.9
ENSMUST00000233533.2 |
Ilrun
|
inflammation and lipid regulator with UBA-like and NBR1-like domains |
| chr10_+_127928622 | 0.37 |
ENSMUST00000219072.2
ENSMUST00000045621.9 ENSMUST00000170054.9 |
Baz2a
|
bromodomain adjacent to zinc finger domain, 2A |
| chr11_+_105072353 | 0.36 |
ENSMUST00000106941.9
|
Tlk2
|
tousled-like kinase 2 (Arabidopsis) |
| chr11_+_96173475 | 0.36 |
ENSMUST00000168043.2
|
Hoxb8
|
homeobox B8 |
| chr17_+_35220252 | 0.36 |
ENSMUST00000174260.8
|
Vars
|
valyl-tRNA synthetase |
| chr15_+_43340609 | 0.36 |
ENSMUST00000022962.8
|
Emc2
|
ER membrane protein complex subunit 2 |
| chr15_+_102314809 | 0.36 |
ENSMUST00000001326.7
|
Sp1
|
trans-acting transcription factor 1 |
| chr1_-_74544946 | 0.36 |
ENSMUST00000044260.11
ENSMUST00000186282.7 |
Usp37
|
ubiquitin specific peptidase 37 |
| chr7_-_24771717 | 0.35 |
ENSMUST00000003468.10
|
Grik5
|
glutamate receptor, ionotropic, kainate 5 (gamma 2) |
| chr8_+_84724130 | 0.34 |
ENSMUST00000095228.5
|
Samd1
|
sterile alpha motif domain containing 1 |
| chr4_+_129083553 | 0.34 |
ENSMUST00000106054.4
|
Yars
|
tyrosyl-tRNA synthetase |
| chr7_-_126303351 | 0.34 |
ENSMUST00000106364.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr17_+_34809132 | 0.34 |
ENSMUST00000173772.2
|
Gpsm3
|
G-protein signalling modulator 3 (AGS3-like, C. elegans) |
| chr11_+_63023893 | 0.34 |
ENSMUST00000108700.2
|
Pmp22
|
peripheral myelin protein 22 |
| chrX_-_46981379 | 0.34 |
ENSMUST00000077569.11
ENSMUST00000101616.9 ENSMUST00000088973.11 |
Smarca1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr11_+_20581952 | 0.34 |
ENSMUST00000109586.3
|
Sertad2
|
SERTA domain containing 2 |
| chr9_-_45895635 | 0.34 |
ENSMUST00000215427.2
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chr11_-_109364424 | 0.33 |
ENSMUST00000070152.12
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr11_+_79883885 | 0.33 |
ENSMUST00000163272.2
ENSMUST00000017692.15 |
Suz12
|
SUZ12 polycomb repressive complex 2 subunit |
| chr8_+_112667328 | 0.33 |
ENSMUST00000034428.8
|
Gabarapl2
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 2 |
| chr10_-_127024641 | 0.33 |
ENSMUST00000218654.2
|
Arhgef25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chr8_+_57908920 | 0.33 |
ENSMUST00000034023.4
|
Scrg1
|
scrapie responsive gene 1 |
| chr13_-_21934675 | 0.33 |
ENSMUST00000102983.2
|
H4c12
|
H4 clustered histone 12 |
| chr7_-_126303689 | 0.32 |
ENSMUST00000135087.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr14_+_45567245 | 0.32 |
ENSMUST00000022380.9
|
Psmc6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr6_+_30047968 | 0.32 |
ENSMUST00000115212.8
ENSMUST00000123194.8 ENSMUST00000115211.8 ENSMUST00000115206.8 ENSMUST00000133928.8 ENSMUST00000115208.8 |
Nrf1
|
nuclear respiratory factor 1 |
| chr11_-_86248395 | 0.32 |
ENSMUST00000043624.9
|
Med13
|
mediator complex subunit 13 |
| chr1_+_86230931 | 0.32 |
ENSMUST00000113306.4
|
B3gnt7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 |
| chr8_+_110432132 | 0.31 |
ENSMUST00000212964.2
ENSMUST00000034163.9 |
Zfp821
|
zinc finger protein 821 |
| chr11_+_105072619 | 0.31 |
ENSMUST00000092537.10
ENSMUST00000015107.13 ENSMUST00000145048.8 |
Tlk2
|
tousled-like kinase 2 (Arabidopsis) |
| chr10_-_7831979 | 0.31 |
ENSMUST00000146444.8
|
Tab2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| chr11_-_69811347 | 0.31 |
ENSMUST00000108610.8
|
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr17_-_24360516 | 0.31 |
ENSMUST00000115411.8
ENSMUST00000115409.9 ENSMUST00000115407.9 ENSMUST00000102927.10 |
Pdpk1
|
3-phosphoinositide dependent protein kinase 1 |
| chr13_-_51947685 | 0.31 |
ENSMUST00000110040.9
ENSMUST00000021900.14 |
Sema4d
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
| chr7_-_89629809 | 0.30 |
ENSMUST00000238792.2
|
Eed
|
embryonic ectoderm development |
| chr10_+_126914755 | 0.30 |
ENSMUST00000039259.7
ENSMUST00000217941.2 |
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr11_-_96868483 | 0.30 |
ENSMUST00000107624.8
|
Sp2
|
Sp2 transcription factor |
| chr15_+_102314578 | 0.30 |
ENSMUST00000170884.8
ENSMUST00000163709.8 |
Sp1
|
trans-acting transcription factor 1 |
| chr7_-_126102579 | 0.30 |
ENSMUST00000040202.15
|
Atxn2l
|
ataxin 2-like |
| chr2_-_30793266 | 0.30 |
ENSMUST00000102852.6
|
Ptges
|
prostaglandin E synthase |
| chr11_+_69792642 | 0.30 |
ENSMUST00000177138.8
ENSMUST00000108617.10 ENSMUST00000177476.8 ENSMUST00000061837.11 |
Neurl4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr17_+_69690314 | 0.29 |
ENSMUST00000062369.14
|
Zbtb14
|
zinc finger and BTB domain containing 14 |
| chr11_-_51891575 | 0.29 |
ENSMUST00000109086.8
|
Ube2b
|
ubiquitin-conjugating enzyme E2B |
| chr5_+_123280250 | 0.29 |
ENSMUST00000174836.8
ENSMUST00000163030.9 |
Setd1b
|
SET domain containing 1B |
| chr10_-_128505096 | 0.29 |
ENSMUST00000238610.2
ENSMUST00000238712.2 |
Ikzf4
|
IKAROS family zinc finger 4 |
| chr11_+_102495189 | 0.29 |
ENSMUST00000057893.7
|
Fzd2
|
frizzled class receptor 2 |
| chrX_+_133657312 | 0.29 |
ENSMUST00000081834.10
ENSMUST00000086880.11 ENSMUST00000086884.5 |
Armcx3
|
armadillo repeat containing, X-linked 3 |
| chr3_+_138231935 | 0.29 |
ENSMUST00000029803.12
|
Eif4e
|
eukaryotic translation initiation factor 4E |
| chr19_-_45800730 | 0.29 |
ENSMUST00000086993.11
|
Kcnip2
|
Kv channel-interacting protein 2 |
| chr8_+_110432210 | 0.29 |
ENSMUST00000212192.2
|
Zfp821
|
zinc finger protein 821 |
| chr7_+_127084283 | 0.28 |
ENSMUST00000048896.8
|
Fbrs
|
fibrosin |
| chr11_-_69649004 | 0.28 |
ENSMUST00000071213.4
|
Polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A |
| chr7_-_126062272 | 0.28 |
ENSMUST00000032974.13
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr7_-_89630141 | 0.28 |
ENSMUST00000238981.2
ENSMUST00000208977.2 ENSMUST00000107234.3 |
Eed
|
embryonic ectoderm development |
| chr10_-_128401773 | 0.28 |
ENSMUST00000026425.13
ENSMUST00000131728.4 |
Pa2g4
|
proliferation-associated 2G4 |
| chr3_+_20111958 | 0.28 |
ENSMUST00000002502.12
|
Hltf
|
helicase-like transcription factor |
| chr4_-_148711211 | 0.28 |
ENSMUST00000186947.7
ENSMUST00000105699.8 |
Tardbp
|
TAR DNA binding protein |
| chr10_-_127177729 | 0.27 |
ENSMUST00000026474.5
ENSMUST00000219671.2 |
Gli1
|
GLI-Kruppel family member GLI1 |
| chr17_+_34808772 | 0.27 |
ENSMUST00000038244.15
|
Gpsm3
|
G-protein signalling modulator 3 (AGS3-like, C. elegans) |
| chr1_-_75195127 | 0.27 |
ENSMUST00000079464.13
|
Tuba4a
|
tubulin, alpha 4A |
| chr8_+_112262729 | 0.27 |
ENSMUST00000172856.8
|
Znrf1
|
zinc and ring finger 1 |
| chr11_+_20493306 | 0.27 |
ENSMUST00000093292.11
|
Sertad2
|
SERTA domain containing 2 |
| chr5_-_107873883 | 0.27 |
ENSMUST00000159263.3
|
Gfi1
|
growth factor independent 1 transcription repressor |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.5 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.5 | 5.9 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.5 | 3.1 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.4 | 1.1 | GO:0034117 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 0.4 | 1.1 | GO:2000474 | cellular response to isoquinoline alkaloid(GO:0071317) regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.3 | 1.5 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.3 | 0.8 | GO:0014737 | positive regulation of muscle atrophy(GO:0014737) |
| 0.3 | 3.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.3 | 1.3 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.2 | 2.7 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.2 | 0.7 | GO:1904826 | regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.2 | 0.6 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) regulation of cGMP-mediated signaling(GO:0010752) |
| 0.2 | 1.2 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.2 | 0.6 | GO:0009955 | adaxial/abaxial pattern specification(GO:0009955) regulation of adaxial/abaxial pattern formation(GO:2000011) |
| 0.2 | 0.6 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) |
| 0.2 | 1.1 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.2 | 0.5 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.2 | 0.5 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) negative regulation of histone phosphorylation(GO:0033128) |
| 0.2 | 1.4 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.2 | 2.0 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.2 | 0.7 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.2 | 3.9 | GO:0043486 | histone exchange(GO:0043486) |
| 0.2 | 0.6 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.2 | 1.0 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.2 | 1.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 1.4 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.2 | 0.5 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.1 | 0.4 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.1 | 0.4 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.1 | 4.8 | GO:0046852 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.1 | 0.5 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 2.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.7 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 1.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 1.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.8 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.1 | 0.4 | GO:0061153 | negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) |
| 0.1 | 0.9 | GO:0051775 | response to redox state(GO:0051775) |
| 0.1 | 0.1 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.1 | 0.7 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.1 | 0.3 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.3 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.1 | 0.3 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.1 | 0.3 | GO:0060032 | notochord regression(GO:0060032) |
| 0.1 | 0.7 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 0.3 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.1 | 1.7 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.1 | GO:0072368 | regulation of lipid transport by negative regulation of transcription from RNA polymerase II promoter(GO:0072368) |
| 0.1 | 0.4 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 0.4 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.1 | 0.5 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.3 | GO:0060220 | camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.1 | 0.2 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 0.8 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.2 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.1 | 0.3 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.6 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.1 | 0.4 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.1 | 0.6 | GO:2000823 | regulation of androgen receptor activity(GO:2000823) |
| 0.1 | 0.7 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 0.2 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.4 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.9 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 0.3 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.1 | 0.6 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.2 | GO:1902689 | negative regulation of NAD metabolic process(GO:1902689) negative regulation of glucose catabolic process to lactate via pyruvate(GO:1904024) |
| 0.1 | 0.2 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.1 | 0.4 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.4 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.1 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.1 | 0.2 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.1 | 0.1 | GO:0060061 | Spemann organizer formation(GO:0060061) canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.1 | 0.5 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 0.3 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.1 | 0.2 | GO:0060466 | activation of meiosis involved in egg activation(GO:0060466) |
| 0.1 | 0.7 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.2 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.1 | 0.4 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 0.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 1.2 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 0.2 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.2 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.2 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.0 | 1.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 0.0 | 0.3 | GO:0097324 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.2 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.1 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.2 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.4 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.0 | 0.5 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.3 | GO:1902661 | regulation of glucose mediated signaling pathway(GO:1902659) positive regulation of glucose mediated signaling pathway(GO:1902661) |
| 0.0 | 0.6 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.9 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.2 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.1 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.0 | 0.4 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.2 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.2 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.0 | 0.1 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0002554 | serotonin secretion by platelet(GO:0002554) |
| 0.0 | 0.2 | GO:1902363 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.4 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.2 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.6 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 0.5 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.2 | GO:0050689 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.2 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.5 | GO:0060982 | coronary artery morphogenesis(GO:0060982) |
| 0.0 | 1.5 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.4 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.0 | 0.3 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.4 | GO:0098953 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.0 | 0.2 | GO:0044334 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.4 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.0 | GO:0060197 | cloacal septation(GO:0060197) |
| 0.0 | 0.6 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.1 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.0 | 0.1 | GO:0021634 | optic nerve formation(GO:0021634) optic chiasma development(GO:0061360) regulation of optic nerve formation(GO:2000595) positive regulation of optic nerve formation(GO:2000597) |
| 0.0 | 0.1 | GO:0015960 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.2 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.1 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.2 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.0 | 0.1 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.0 | 0.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.0 | 0.1 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.0 | 0.2 | GO:0050757 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.3 | GO:0034088 | maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.0 | GO:1904753 | negative regulation of vascular associated smooth muscle cell migration(GO:1904753) |
| 0.0 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.8 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 0.4 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.2 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.2 | GO:1904948 | midbrain dopaminergic neuron differentiation(GO:1904948) |
| 0.0 | 0.5 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:1905049 | negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 | 0.1 | GO:0100012 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) |
| 0.0 | 0.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0045358 | negative regulation of interferon-beta biosynthetic process(GO:0045358) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.3 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.0 | 0.7 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.1 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.1 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.0 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.1 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0072086 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) specification of loop of Henle identity(GO:0072086) |
| 0.0 | 2.1 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.0 | GO:0021750 | cerebellar molecular layer development(GO:0021679) vestibular nucleus development(GO:0021750) |
| 0.0 | 0.1 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.2 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.2 | GO:1904871 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.2 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.5 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.2 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.2 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.4 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.2 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.1 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.1 | GO:0097116 | protein complex assembly involved in synapse maturation(GO:0090126) gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.4 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.4 | GO:0060749 | mammary gland alveolus development(GO:0060749) mammary gland lobule development(GO:0061377) |
| 0.0 | 0.1 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.1 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.5 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.3 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.2 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.2 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) |
| 0.0 | 0.3 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.0 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.0 | 0.3 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.5 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.4 | 3.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.4 | 1.1 | GO:0090537 | CERF complex(GO:0090537) |
| 0.3 | 1.3 | GO:0001740 | Barr body(GO:0001740) |
| 0.3 | 0.9 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.2 | 0.8 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.2 | 2.0 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 1.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 1.0 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.4 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.6 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 2.0 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.8 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.8 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 1.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.1 | 0.3 | GO:0042025 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.1 | 1.0 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 0.5 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 0.2 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 0.5 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.4 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.2 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 0.1 | 0.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.2 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.2 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.1 | 0.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.7 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.3 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.4 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 2.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.1 | GO:0032998 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.0 | 1.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.3 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 2.7 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 1.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 2.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.6 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.6 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.2 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.0 | 0.7 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.2 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.3 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.4 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.9 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 2.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.6 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.2 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.1 | GO:1990696 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.0 | 0.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.6 | GO:0032993 | protein-DNA complex(GO:0032993) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.0 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.3 | 3.1 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 0.7 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 0.2 | 1.4 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.2 | 2.7 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 2.9 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 0.9 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 3.5 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.2 | 0.5 | GO:0090556 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.2 | 0.9 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.2 | 0.6 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.8 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 1.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 1.0 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.4 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.1 | 1.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.1 | 1.5 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 1.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.5 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.3 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 0.8 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 2.7 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.7 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.4 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.1 | 0.2 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.1 | 0.8 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.3 | GO:0050220 | prostaglandin-D synthase activity(GO:0004667) prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 4.2 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 0.7 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 1.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.4 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.1 | 0.4 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 0.4 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 0.4 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.6 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 0.6 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 1.0 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 1.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 1.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 6.7 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 2.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.3 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.5 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.5 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 1.0 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.7 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.3 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.2 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 1.6 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.8 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 3.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.7 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.9 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.3 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.3 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.4 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 1.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.7 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.4 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 1.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 3.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.8 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 2.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.7 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.9 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 2.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 2.2 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.8 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.0 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 4.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 3.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.5 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 0.6 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 1.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 0.2 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 3.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.8 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.9 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.8 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.0 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.7 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.0 | 0.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 1.2 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.2 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.9 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 1.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.2 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.0 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.5 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |