Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
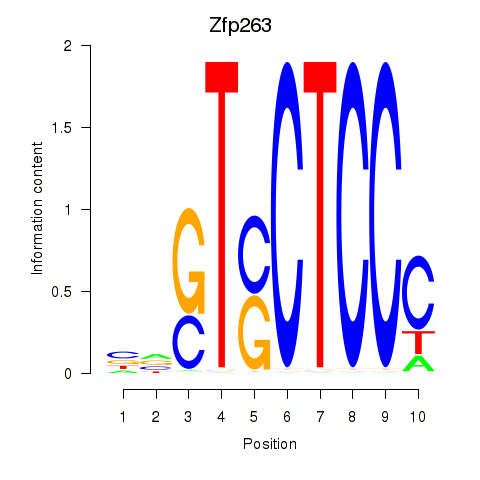
Results for Zfp263
Z-value: 2.22
Transcription factors associated with Zfp263
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp263
|
ENSMUSG00000022529.12 | Zfp263 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp263 | mm39_v1_chr16_+_3561952_3562023 | -0.59 | 1.6e-04 | Click! |
Activity profile of Zfp263 motif
Sorted Z-values of Zfp263 motif
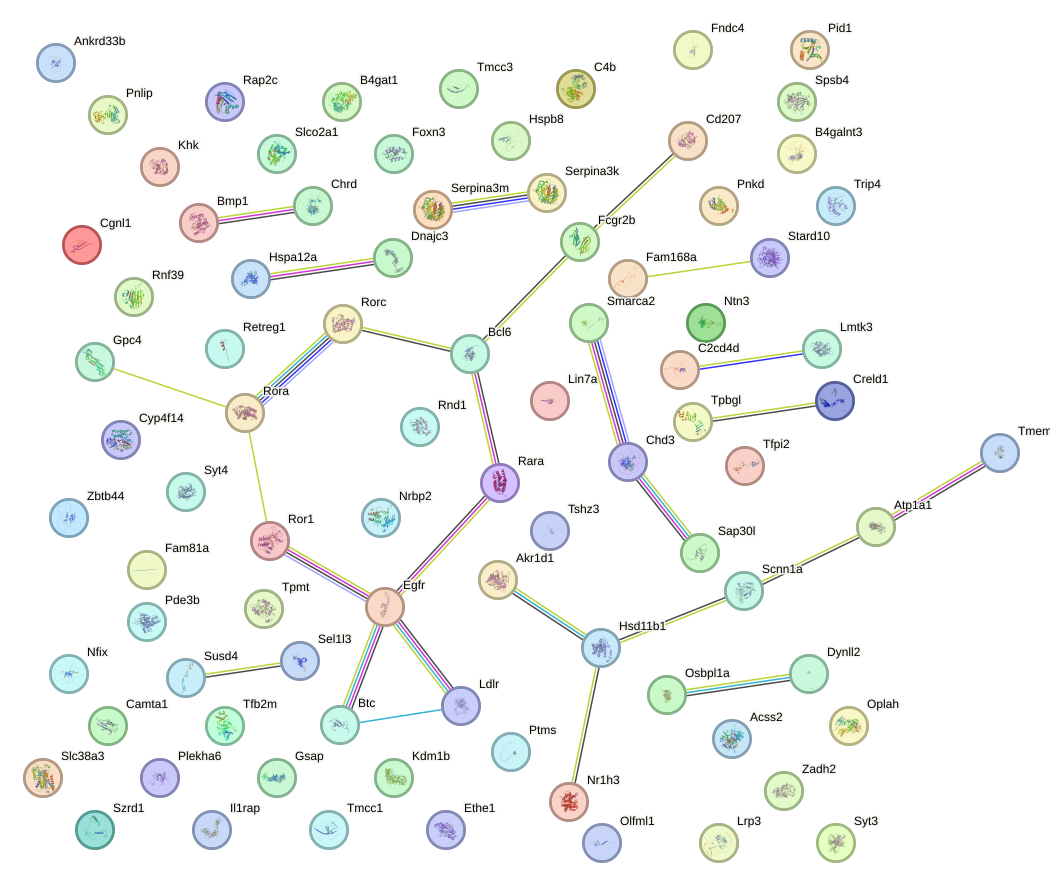
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp263
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_104304631 | 12.42 |
ENSMUST00000043058.5
ENSMUST00000101078.12 |
Serpina3k
Serpina3m
|
serine (or cysteine) peptidase inhibitor, clade A, member 3K serine (or cysteine) peptidase inhibitor, clade A, member 3M |
| chr2_+_58457370 | 10.98 |
ENSMUST00000071543.12
|
Upp2
|
uridine phosphorylase 2 |
| chr5_+_31078775 | 10.30 |
ENSMUST00000201621.4
|
Khk
|
ketohexokinase |
| chr1_-_192946359 | 8.67 |
ENSMUST00000161737.8
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr5_+_31078911 | 8.50 |
ENSMUST00000201571.4
|
Khk
|
ketohexokinase |
| chr11_+_16702203 | 7.94 |
ENSMUST00000102884.10
ENSMUST00000020329.13 |
Egfr
|
epidermal growth factor receptor |
| chr17_-_33136277 | 7.68 |
ENSMUST00000234538.2
ENSMUST00000235058.2 ENSMUST00000234759.2 ENSMUST00000179434.8 ENSMUST00000234797.2 |
Cyp4f14
|
cytochrome P450, family 4, subfamily f, polypeptide 14 |
| chr17_-_33136021 | 6.80 |
ENSMUST00000054174.9
|
Cyp4f14
|
cytochrome P450, family 4, subfamily f, polypeptide 14 |
| chr5_+_31079177 | 6.21 |
ENSMUST00000031053.15
ENSMUST00000202752.2 |
Khk
|
ketohexokinase |
| chr11_-_74816484 | 5.62 |
ENSMUST00000138612.2
ENSMUST00000123855.8 ENSMUST00000128556.8 ENSMUST00000108448.8 ENSMUST00000108447.8 ENSMUST00000065211.9 |
Srr
|
serine racemase |
| chr19_+_58658779 | 5.14 |
ENSMUST00000057270.9
|
Pnlip
|
pancreatic lipase |
| chr17_-_34962823 | 5.10 |
ENSMUST00000069507.9
|
C4b
|
complement component 4B (Chido blood group) |
| chr6_-_124840824 | 5.02 |
ENSMUST00000046893.10
ENSMUST00000204667.2 |
Gpr162
|
G protein-coupled receptor 162 |
| chr7_+_44033520 | 4.90 |
ENSMUST00000118962.8
ENSMUST00000118831.8 |
Syt3
|
synaptotagmin III |
| chr11_-_74816750 | 4.71 |
ENSMUST00000121738.8
|
Srr
|
serine racemase |
| chr16_+_26400454 | 4.63 |
ENSMUST00000096129.9
ENSMUST00000166294.9 ENSMUST00000174202.8 ENSMUST00000023156.13 |
Il1rap
|
interleukin 1 receptor accessory protein |
| chr19_+_58658838 | 4.54 |
ENSMUST00000238108.2
|
Pnlip
|
pancreatic lipase |
| chr15_-_76191301 | 4.52 |
ENSMUST00000171340.9
ENSMUST00000023222.13 ENSMUST00000164189.2 |
Oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr6_+_37507108 | 4.33 |
ENSMUST00000040987.11
|
Akr1d1
|
aldo-keto reductase family 1, member D1 |
| chr1_-_170803680 | 3.66 |
ENSMUST00000027966.14
ENSMUST00000081103.12 ENSMUST00000159688.2 |
Fcgr2b
|
Fc receptor, IgG, low affinity IIb |
| chr6_-_124894902 | 3.65 |
ENSMUST00000032216.7
|
Ptms
|
parathymosin |
| chr15_-_98575332 | 3.62 |
ENSMUST00000120997.2
ENSMUST00000109149.9 ENSMUST00000003451.11 |
Rnd1
|
Rho family GTPase 1 |
| chr17_-_24428351 | 3.46 |
ENSMUST00000024931.6
|
Ntn3
|
netrin 3 |
| chr6_+_125298372 | 3.38 |
ENSMUST00000176442.8
ENSMUST00000177329.2 |
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr13_-_47196633 | 3.36 |
ENSMUST00000021806.11
ENSMUST00000136864.8 |
Tpmt
|
thiopurine methyltransferase |
| chr11_-_87878301 | 3.28 |
ENSMUST00000020775.9
|
Dynll2
|
dynein light chain LC8-type 2 |
| chr7_+_107166653 | 3.27 |
ENSMUST00000120990.2
|
Olfml1
|
olfactomedin-like 1 |
| chr18_-_12995261 | 3.24 |
ENSMUST00000234427.2
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr16_-_23807602 | 3.24 |
ENSMUST00000023151.6
|
Bcl6
|
B cell leukemia/lymphoma 6 |
| chr13_-_47196592 | 3.19 |
ENSMUST00000110118.8
|
Tpmt
|
thiopurine methyltransferase |
| chr8_-_85526653 | 3.12 |
ENSMUST00000126806.2
ENSMUST00000076715.13 |
Nfix
|
nuclear factor I/X |
| chr3_+_94280101 | 3.10 |
ENSMUST00000029795.10
|
Rorc
|
RAR-related orphan receptor gamma |
| chr7_+_119360141 | 3.06 |
ENSMUST00000106528.8
ENSMUST00000106527.8 ENSMUST00000063770.10 ENSMUST00000106529.8 |
Acsm3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr12_-_99359265 | 3.02 |
ENSMUST00000177451.8
|
Foxn3
|
forkhead box N3 |
| chr13_-_47196607 | 3.00 |
ENSMUST00000124948.2
|
Tpmt
|
thiopurine methyltransferase |
| chr9_-_107544573 | 2.91 |
ENSMUST00000010208.14
ENSMUST00000193932.6 |
Slc38a3
|
solute carrier family 38, member 3 |
| chr16_+_20551853 | 2.90 |
ENSMUST00000115423.8
ENSMUST00000007171.13 ENSMUST00000232646.2 |
Chrd
|
chordin |
| chr17_+_37253802 | 2.90 |
ENSMUST00000040498.12
|
Rnf39
|
ring finger protein 39 |
| chr7_-_34914675 | 2.89 |
ENSMUST00000118444.3
ENSMUST00000122409.8 |
Lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr19_+_5088854 | 2.88 |
ENSMUST00000053705.8
ENSMUST00000235776.2 |
B4gat1
|
beta-1,4-glucuronyltransferase 1 |
| chrX_-_13712746 | 2.87 |
ENSMUST00000115436.9
ENSMUST00000033321.11 ENSMUST00000115438.10 |
Cask
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr5_-_116560916 | 2.85 |
ENSMUST00000036991.5
|
Hspb8
|
heat shock protein 8 |
| chr2_+_155360015 | 2.83 |
ENSMUST00000103142.12
|
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr7_+_107166925 | 2.82 |
ENSMUST00000239087.2
|
Olfml1
|
olfactomedin-like 1 |
| chr2_+_155359868 | 2.77 |
ENSMUST00000029135.15
ENSMUST00000065973.9 |
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr5_+_21391282 | 2.76 |
ENSMUST00000036031.13
ENSMUST00000198937.2 |
Gsap
|
gamma-secretase activating protein |
| chr14_+_119175328 | 2.75 |
ENSMUST00000022734.9
|
Dnajc3
|
DnaJ heat shock protein family (Hsp40) member C3 |
| chr1_-_170804116 | 2.66 |
ENSMUST00000159969.8
|
Fcgr2b
|
Fc receptor, IgG, low affinity IIb |
| chr2_-_147887810 | 2.65 |
ENSMUST00000109964.8
|
Foxa2
|
forkhead box A2 |
| chr7_+_24286968 | 2.63 |
ENSMUST00000077191.7
|
Ethe1
|
ethylmalonic encephalopathy 1 |
| chrX_-_51254129 | 2.62 |
ENSMUST00000033450.3
|
Gpc4
|
glypican 4 |
| chr5_-_31453206 | 2.56 |
ENSMUST00000041266.11
ENSMUST00000172435.8 ENSMUST00000201417.2 |
Fndc4
|
fibronectin type III domain containing 4 |
| chr8_-_85526972 | 2.55 |
ENSMUST00000099070.10
|
Nfix
|
nuclear factor I/X |
| chr15_-_31367872 | 2.52 |
ENSMUST00000123325.9
|
Ankrd33b
|
ankyrin repeat domain 33B |
| chr18_-_12995681 | 2.50 |
ENSMUST00000121808.8
ENSMUST00000118313.8 |
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr7_+_114014509 | 2.47 |
ENSMUST00000032909.9
|
Pde3b
|
phosphodiesterase 3B, cGMP-inhibited |
| chr15_-_75963446 | 2.43 |
ENSMUST00000228366.3
|
Nrbp2
|
nuclear receptor binding protein 2 |
| chr7_-_99276310 | 2.42 |
ENSMUST00000178124.3
|
Tpbgl
|
trophoblast glycoprotein-like |
| chr6_-_120271520 | 2.41 |
ENSMUST00000057283.8
ENSMUST00000212457.2 |
B4galnt3
|
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
| chr15_+_25940912 | 2.39 |
ENSMUST00000226438.2
|
Retreg1
|
reticulophagy regulator 1 |
| chr9_-_79700660 | 2.38 |
ENSMUST00000034878.12
|
Tmem30a
|
transmembrane protein 30A |
| chr6_-_119521243 | 2.36 |
ENSMUST00000119369.2
ENSMUST00000178696.8 |
Wnt5b
|
wingless-type MMTV integration site family, member 5B |
| chr6_-_83654789 | 2.34 |
ENSMUST00000037882.8
|
Cd207
|
CD207 antigen |
| chr7_+_45434755 | 2.33 |
ENSMUST00000233503.2
ENSMUST00000120005.10 ENSMUST00000211609.2 |
Lmtk3
|
lemur tyrosine kinase 3 |
| chr7_+_100355798 | 2.33 |
ENSMUST00000107042.9
ENSMUST00000207564.2 ENSMUST00000049053.9 |
Fam168a
|
family with sequence similarity 168, member A |
| chr3_+_94269745 | 2.32 |
ENSMUST00000169433.3
|
C2cd4d
|
C2 calcium-dependent domain containing 4D |
| chr4_-_151946155 | 2.32 |
ENSMUST00000049790.14
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr19_+_26582450 | 2.31 |
ENSMUST00000176769.9
ENSMUST00000208163.2 ENSMUST00000025862.15 ENSMUST00000176030.8 |
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr11_-_69260203 | 2.30 |
ENSMUST00000092971.13
ENSMUST00000108661.8 |
Chd3
|
chromodomain helicase DNA binding protein 3 |
| chr7_+_36397426 | 2.29 |
ENSMUST00000021641.8
|
Tshz3
|
teashirt zinc finger family member 3 |
| chr5_-_91550853 | 2.29 |
ENSMUST00000121044.6
|
Btc
|
betacellulin, epidermal growth factor family member |
| chr4_-_151946219 | 2.28 |
ENSMUST00000097774.9
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr15_+_25940781 | 2.28 |
ENSMUST00000227275.2
|
Retreg1
|
reticulophagy regulator 1 |
| chr9_+_102885156 | 2.25 |
ENSMUST00000035148.13
|
Slco2a1
|
solute carrier organic anion transporter family, member 2a1 |
| chr6_+_125298296 | 2.24 |
ENSMUST00000081440.14
|
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr9_+_21634918 | 2.23 |
ENSMUST00000213114.2
|
Ldlr
|
low density lipoprotein receptor |
| chr11_+_57692399 | 2.22 |
ENSMUST00000020826.6
|
Sap30l
|
SAP30-like |
| chr1_+_74324089 | 2.21 |
ENSMUST00000113805.8
ENSMUST00000027370.13 ENSMUST00000087226.11 |
Pnkd
|
paroxysmal nonkinesiogenic dyskinesia |
| chr2_-_91025208 | 2.21 |
ENSMUST00000111355.8
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr6_+_113460258 | 2.21 |
ENSMUST00000032422.6
|
Creld1
|
cysteine-rich with EGF-like domains 1 |
| chr6_-_3968365 | 2.20 |
ENSMUST00000031674.11
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr9_-_65792306 | 2.19 |
ENSMUST00000122410.8
ENSMUST00000117083.2 |
Trip4
|
thyroid hormone receptor interactor 4 |
| chr9_-_71678814 | 2.16 |
ENSMUST00000122065.2
ENSMUST00000121322.8 ENSMUST00000072899.9 |
Cgnl1
|
cingulin-like 1 |
| chr18_-_12995395 | 2.14 |
ENSMUST00000121888.8
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr15_-_31367668 | 2.12 |
ENSMUST00000110410.10
ENSMUST00000076942.5 |
Ankrd33b
|
ankyrin repeat domain 33B |
| chr2_-_91025441 | 2.12 |
ENSMUST00000002177.9
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr15_+_25940931 | 2.12 |
ENSMUST00000110438.3
|
Retreg1
|
reticulophagy regulator 1 |
| chr7_+_100970435 | 2.12 |
ENSMUST00000210192.2
ENSMUST00000172630.8 |
Stard10
|
START domain containing 10 |
| chr1_-_179373826 | 2.09 |
ENSMUST00000027769.6
|
Tfb2m
|
transcription factor B2, mitochondrial |
| chr1_+_182591771 | 2.07 |
ENSMUST00000193660.6
|
Susd4
|
sushi domain containing 4 |
| chr1_-_84262274 | 2.06 |
ENSMUST00000177458.2
ENSMUST00000168574.9 |
Pid1
|
phosphotyrosine interaction domain containing 1 |
| chrX_-_50106844 | 2.04 |
ENSMUST00000053593.8
|
Rap2c
|
RAP2C, member of RAS oncogene family |
| chr2_-_91025380 | 2.02 |
ENSMUST00000111356.8
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr11_+_70591299 | 2.01 |
ENSMUST00000152618.9
ENSMUST00000102554.8 ENSMUST00000094499.11 ENSMUST00000072187.12 ENSMUST00000137119.3 |
Kif1c
|
kinesin family member 1C |
| chr7_+_100970287 | 2.01 |
ENSMUST00000032927.14
|
Stard10
|
START domain containing 10 |
| chr3_-_89294430 | 1.97 |
ENSMUST00000107433.8
|
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr18_-_12995913 | 1.97 |
ENSMUST00000121774.8
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr9_-_70048766 | 1.97 |
ENSMUST00000034749.16
|
Fam81a
|
family with sequence similarity 81, member A |
| chr4_+_99952988 | 1.96 |
ENSMUST00000039630.6
|
Ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr3_-_101511971 | 1.96 |
ENSMUST00000036493.8
|
Atp1a1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
| chr8_-_85500998 | 1.95 |
ENSMUST00000109762.8
|
Nfix
|
nuclear factor I/X |
| chr9_+_21634779 | 1.94 |
ENSMUST00000034713.9
|
Ldlr
|
low density lipoprotein receptor |
| chr9_+_30941924 | 1.91 |
ENSMUST00000216649.2
ENSMUST00000115222.10 |
Zbtb44
|
zinc finger and BTB domain containing 44 |
| chr18_+_84106188 | 1.91 |
ENSMUST00000060223.4
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2 |
| chr3_-_146321341 | 1.88 |
ENSMUST00000200633.2
|
Dnase2b
|
deoxyribonuclease II beta |
| chr6_-_116084810 | 1.87 |
ENSMUST00000204353.3
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr9_-_96900876 | 1.85 |
ENSMUST00000055433.5
|
Spsb4
|
splA/ryanodine receptor domain and SOCS box containing 4 |
| chr19_-_58849407 | 1.84 |
ENSMUST00000066285.6
|
Hspa12a
|
heat shock protein 12A |
| chr11_+_98851238 | 1.83 |
ENSMUST00000107473.3
|
Rara
|
retinoic acid receptor, alpha |
| chr10_+_107107477 | 1.81 |
ENSMUST00000020057.16
|
Lin7a
|
lin-7 homolog A (C. elegans) |
| chr1_+_182591425 | 1.80 |
ENSMUST00000155229.7
ENSMUST00000153348.8 |
Susd4
|
sushi domain containing 4 |
| chr7_+_100970910 | 1.77 |
ENSMUST00000174291.8
ENSMUST00000167888.9 ENSMUST00000172662.2 |
Stard10
|
START domain containing 10 |
| chr13_+_47196975 | 1.76 |
ENSMUST00000037025.16
ENSMUST00000143868.2 |
Kdm1b
|
lysine (K)-specific demethylase 1B |
| chr5_-_53370761 | 1.76 |
ENSMUST00000031090.8
|
Sel1l3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr10_+_107107558 | 1.76 |
ENSMUST00000105280.5
|
Lin7a
|
lin-7 homolog A (C. elegans) |
| chr3_+_40904253 | 1.74 |
ENSMUST00000048490.13
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr1_-_155912159 | 1.73 |
ENSMUST00000097527.10
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr4_-_140867038 | 1.73 |
ENSMUST00000148204.8
ENSMUST00000102487.4 |
Szrd1
|
SUZ RNA binding domain containing 1 |
| chr1_-_84262144 | 1.73 |
ENSMUST00000176720.2
|
Pid1
|
phosphotyrosine interaction domain containing 1 |
| chr14_-_70757601 | 1.71 |
ENSMUST00000022693.9
|
Bmp1
|
bone morphogenetic protein 1 |
| chr1_+_133109059 | 1.69 |
ENSMUST00000187285.7
|
Plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr3_+_40905066 | 1.68 |
ENSMUST00000191805.7
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr11_-_96714950 | 1.68 |
ENSMUST00000169828.8
ENSMUST00000126949.8 |
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1 |
| chr16_-_59421342 | 1.68 |
ENSMUST00000172910.3
|
Crybg3
|
beta-gamma crystallin domain containing 3 |
| chr7_-_127423641 | 1.67 |
ENSMUST00000106267.5
|
Stx1b
|
syntaxin 1B |
| chr11_-_96714813 | 1.67 |
ENSMUST00000142065.2
ENSMUST00000167110.8 |
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1 |
| chr7_-_80051455 | 1.66 |
ENSMUST00000120753.3
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr7_+_100355910 | 1.65 |
ENSMUST00000207875.2
ENSMUST00000208013.2 |
Fam168a
|
family with sequence similarity 168, member A |
| chr6_-_29507944 | 1.64 |
ENSMUST00000101614.10
ENSMUST00000078112.13 |
Kcp
|
kielin/chordin-like protein |
| chr8_+_124380639 | 1.64 |
ENSMUST00000045487.4
|
Rhou
|
ras homolog family member U |
| chr12_+_104372962 | 1.63 |
ENSMUST00000021506.6
|
Serpina3n
|
serine (or cysteine) peptidase inhibitor, clade A, member 3N |
| chr9_-_79700789 | 1.62 |
ENSMUST00000120690.2
|
Tmem30a
|
transmembrane protein 30A |
| chrX_+_141009756 | 1.62 |
ENSMUST00000112916.9
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr17_-_12726591 | 1.62 |
ENSMUST00000024595.4
|
Slc22a3
|
solute carrier family 22 (organic cation transporter), member 3 |
| chr4_-_129121676 | 1.62 |
ENSMUST00000106051.8
|
C77080
|
expressed sequence C77080 |
| chr6_+_88701810 | 1.61 |
ENSMUST00000089449.5
|
Mgll
|
monoglyceride lipase |
| chr2_-_91025492 | 1.61 |
ENSMUST00000111354.2
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr6_+_88701394 | 1.61 |
ENSMUST00000113585.9
|
Mgll
|
monoglyceride lipase |
| chr6_-_148732946 | 1.59 |
ENSMUST00000048418.14
|
Ipo8
|
importin 8 |
| chr15_-_53209513 | 1.59 |
ENSMUST00000077273.9
|
Ext1
|
exostosin glycosyltransferase 1 |
| chr9_+_44018583 | 1.59 |
ENSMUST00000152956.8
ENSMUST00000114815.3 ENSMUST00000206295.2 ENSMUST00000206769.2 ENSMUST00000205500.2 |
C1qtnf5
|
C1q and tumor necrosis factor related protein 5 |
| chr3_+_40905216 | 1.55 |
ENSMUST00000191872.6
ENSMUST00000200432.2 |
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr1_+_153300874 | 1.55 |
ENSMUST00000042373.12
|
Shcbp1l
|
Shc SH2-domain binding protein 1-like |
| chr1_+_155911879 | 1.55 |
ENSMUST00000128941.8
|
Tor1aip2
|
torsin A interacting protein 2 |
| chr12_+_105302853 | 1.53 |
ENSMUST00000180458.9
|
Tunar
|
Tcl1 upstream neural differentiation associated RNA |
| chr7_+_24310171 | 1.53 |
ENSMUST00000206422.2
|
Phldb3
|
pleckstrin homology like domain, family B, member 3 |
| chrX_+_57075981 | 1.52 |
ENSMUST00000088627.11
|
Zic3
|
zinc finger protein of the cerebellum 3 |
| chr18_-_60881405 | 1.51 |
ENSMUST00000237070.2
|
Ndst1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr6_-_148732893 | 1.51 |
ENSMUST00000145960.2
|
Ipo8
|
importin 8 |
| chr5_+_111478608 | 1.50 |
ENSMUST00000086635.9
ENSMUST00000200298.2 |
Pitpnb
|
phosphatidylinositol transfer protein, beta |
| chr18_-_60881679 | 1.50 |
ENSMUST00000237783.2
|
Ndst1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr12_-_104120105 | 1.49 |
ENSMUST00000085050.4
|
Serpina3c
|
serine (or cysteine) peptidase inhibitor, clade A, member 3C |
| chr11_-_6469494 | 1.48 |
ENSMUST00000134489.2
|
Myo1g
|
myosin IG |
| chr3_-_95646856 | 1.47 |
ENSMUST00000153026.8
ENSMUST00000123143.8 ENSMUST00000137912.8 ENSMUST00000029753.14 ENSMUST00000131376.8 ENSMUST00000117507.10 ENSMUST00000128885.8 ENSMUST00000147217.2 |
Ecm1
|
extracellular matrix protein 1 |
| chr14_-_34224620 | 1.46 |
ENSMUST00000049005.15
|
Bmpr1a
|
bone morphogenetic protein receptor, type 1A |
| chr7_-_103320398 | 1.46 |
ENSMUST00000062144.4
|
Olfr624
|
olfactory receptor 624 |
| chr4_+_104770653 | 1.45 |
ENSMUST00000106803.9
ENSMUST00000106804.2 |
Fyb2
|
FYN binding protein 2 |
| chr4_-_151946124 | 1.44 |
ENSMUST00000169423.9
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr1_+_23801084 | 1.44 |
ENSMUST00000140583.2
|
B3gat2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr6_-_48422307 | 1.43 |
ENSMUST00000114563.8
ENSMUST00000114558.8 ENSMUST00000101443.10 |
Zfp467
|
zinc finger protein 467 |
| chr11_+_99755302 | 1.41 |
ENSMUST00000092694.4
|
Gm11559
|
predicted gene 11559 |
| chr2_-_84605732 | 1.41 |
ENSMUST00000023994.10
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr18_+_36098090 | 1.40 |
ENSMUST00000176873.8
ENSMUST00000177432.8 ENSMUST00000175734.2 |
Psd2
|
pleckstrin and Sec7 domain containing 2 |
| chr2_+_163389068 | 1.40 |
ENSMUST00000109411.8
ENSMUST00000018094.13 |
Hnf4a
|
hepatic nuclear factor 4, alpha |
| chr9_+_44018551 | 1.40 |
ENSMUST00000114821.9
ENSMUST00000114818.9 |
C1qtnf5
|
C1q and tumor necrosis factor related protein 5 |
| chr15_+_102011352 | 1.39 |
ENSMUST00000169627.9
|
Tns2
|
tensin 2 |
| chr6_-_48422447 | 1.38 |
ENSMUST00000114564.8
|
Zfp467
|
zinc finger protein 467 |
| chr15_+_102011415 | 1.38 |
ENSMUST00000046144.10
|
Tns2
|
tensin 2 |
| chr18_+_31892933 | 1.36 |
ENSMUST00000115808.4
|
Ammecr1l
|
AMME chromosomal region gene 1-like |
| chr11_+_103007054 | 1.36 |
ENSMUST00000053063.7
|
Hexim1
|
hexamethylene bis-acetamide inducible 1 |
| chr6_+_88701444 | 1.32 |
ENSMUST00000113582.8
|
Mgll
|
monoglyceride lipase |
| chr16_+_24212284 | 1.32 |
ENSMUST00000038053.14
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr18_-_75830595 | 1.31 |
ENSMUST00000165559.3
|
Ctif
|
CBP80/20-dependent translation initiation factor |
| chr1_-_155912216 | 1.31 |
ENSMUST00000027738.14
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr6_+_134807097 | 1.30 |
ENSMUST00000046303.12
|
Crebl2
|
cAMP responsive element binding protein-like 2 |
| chr6_-_84565613 | 1.28 |
ENSMUST00000204146.3
|
Cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr16_+_31482949 | 1.28 |
ENSMUST00000023454.12
|
Dlg1
|
discs large MAGUK scaffold protein 1 |
| chr13_+_58550499 | 1.28 |
ENSMUST00000225815.2
|
Rmi1
|
RecQ mediated genome instability 1 |
| chr13_-_54759145 | 1.28 |
ENSMUST00000091609.11
|
Cltb
|
clathrin, light polypeptide (Lcb) |
| chr11_+_112673041 | 1.26 |
ENSMUST00000000579.3
|
Sox9
|
SRY (sex determining region Y)-box 9 |
| chr13_+_58550046 | 1.26 |
ENSMUST00000225828.2
ENSMUST00000042450.10 |
Rmi1
|
RecQ mediated genome instability 1 |
| chr11_-_43727071 | 1.26 |
ENSMUST00000167574.2
|
Adra1b
|
adrenergic receptor, alpha 1b |
| chr4_-_133066594 | 1.26 |
ENSMUST00000043305.14
|
Wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr2_+_90948481 | 1.26 |
ENSMUST00000137942.8
ENSMUST00000111430.10 ENSMUST00000169776.2 |
Mybpc3
|
myosin binding protein C, cardiac |
| chr4_+_95855442 | 1.24 |
ENSMUST00000030306.14
|
Hook1
|
hook microtubule tethering protein 1 |
| chr9_-_50639230 | 1.24 |
ENSMUST00000118707.2
ENSMUST00000034566.15 |
Dixdc1
|
DIX domain containing 1 |
| chr11_-_78313043 | 1.24 |
ENSMUST00000001122.6
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr15_-_76084035 | 1.23 |
ENSMUST00000054449.14
ENSMUST00000169714.8 ENSMUST00000165453.8 |
Plec
|
plectin |
| chr19_-_7194912 | 1.22 |
ENSMUST00000039758.6
|
Cox8a
|
cytochrome c oxidase subunit 8A |
| chr12_+_72807985 | 1.22 |
ENSMUST00000021514.10
|
Ppm1a
|
protein phosphatase 1A, magnesium dependent, alpha isoform |
| chr11_-_107685383 | 1.21 |
ENSMUST00000021066.4
|
Cacng4
|
calcium channel, voltage-dependent, gamma subunit 4 |
| chr6_+_88701470 | 1.20 |
ENSMUST00000113581.8
|
Mgll
|
monoglyceride lipase |
| chr15_+_34082805 | 1.19 |
ENSMUST00000022865.17
|
Mtdh
|
metadherin |
| chr5_-_50216249 | 1.18 |
ENSMUST00000030971.7
|
Adgra3
|
adhesion G protein-coupled receptor A3 |
| chr1_+_165288606 | 1.18 |
ENSMUST00000027853.6
|
Mpc2
|
mitochondrial pyruvate carrier 2 |
| chr17_+_81251997 | 1.17 |
ENSMUST00000025092.5
|
Tmem178
|
transmembrane protein 178 |
| chr12_+_24622274 | 1.15 |
ENSMUST00000085553.13
|
Grhl1
|
grainyhead like transcription factor 1 |
| chr11_+_97554192 | 1.15 |
ENSMUST00000044730.12
|
Mllt6
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 6 |
| chr14_+_30673334 | 1.15 |
ENSMUST00000226551.2
ENSMUST00000228328.2 |
Nek4
|
NIMA (never in mitosis gene a)-related expressed kinase 4 |
| chr4_-_138095277 | 1.14 |
ENSMUST00000030535.4
|
Cda
|
cytidine deaminase |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 25.0 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 2.3 | 6.9 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 2.1 | 10.3 | GO:0070178 | D-serine metabolic process(GO:0070178) |
| 2.0 | 8.0 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 1.9 | 7.8 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 1.9 | 5.6 | GO:0019427 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 1.4 | 4.3 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 1.3 | 7.9 | GO:0070459 | prolactin secretion(GO:0070459) regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 1.0 | 4.2 | GO:1905167 | regulation of phosphatidylcholine catabolic process(GO:0010899) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 1.0 | 4.0 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 1.0 | 3.0 | GO:1900062 | regulation of replicative cell aging(GO:1900062) |
| 1.0 | 2.9 | GO:0006867 | asparagine transport(GO:0006867) positive regulation of glutamine transport(GO:2000487) |
| 1.0 | 3.9 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 1.0 | 5.7 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.9 | 2.6 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.8 | 3.2 | GO:0043380 | regulation of memory T cell differentiation(GO:0043380) |
| 0.8 | 7.1 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.8 | 2.3 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.8 | 3.8 | GO:2001274 | positive regulation of fat cell proliferation(GO:0070346) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.7 | 2.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.6 | 9.7 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.6 | 1.8 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.6 | 3.6 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.6 | 1.8 | GO:1904826 | regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.6 | 5.9 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.6 | 2.8 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.5 | 1.1 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.5 | 0.5 | GO:0060844 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel endothelial cell fate specification(GO:0097101) |
| 0.5 | 2.6 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.5 | 2.1 | GO:0071503 | response to heparin(GO:0071503) cellular response to heparin(GO:0071504) |
| 0.5 | 4.6 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.5 | 2.0 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.5 | 1.5 | GO:1904414 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) positive regulation of cardiac ventricle development(GO:1904414) |
| 0.5 | 2.9 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.5 | 2.4 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.5 | 1.4 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.5 | 1.4 | GO:1902569 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 0.5 | 1.4 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 0.4 | 0.9 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.4 | 1.8 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.4 | 2.5 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.4 | 8.9 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.4 | 1.3 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.4 | 1.7 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.4 | 1.2 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.4 | 4.7 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.4 | 1.2 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.4 | 1.1 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.3 | 1.7 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.3 | 2.4 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.3 | 2.3 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.3 | 1.3 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.3 | 1.3 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.3 | 1.3 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.3 | 0.9 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.3 | 4.0 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.3 | 3.3 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.3 | 3.0 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 0.3 | 0.9 | GO:2000536 | negative regulation of entry of bacterium into host cell(GO:2000536) |
| 0.3 | 0.9 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.3 | 3.1 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.3 | 3.0 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.3 | 0.8 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.3 | 1.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.3 | 4.9 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.3 | 1.0 | GO:0007522 | visceral muscle development(GO:0007522) |
| 0.3 | 0.8 | GO:0009955 | adaxial/abaxial pattern specification(GO:0009955) regulation of adaxial/abaxial pattern formation(GO:2000011) |
| 0.3 | 0.3 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.2 | 0.7 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.2 | 1.7 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.2 | 0.7 | GO:0060715 | Spemann organizer formation(GO:0060061) syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.2 | 1.0 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 | 1.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.2 | 0.7 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.2 | 0.5 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.2 | 1.6 | GO:0048208 | trophectodermal cellular morphogenesis(GO:0001831) vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.2 | 1.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 0.7 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.2 | 2.2 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.2 | 1.8 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.2 | 2.6 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.2 | 0.9 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.2 | 0.7 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.2 | 0.9 | GO:0098961 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.2 | 1.7 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.2 | 1.3 | GO:1902811 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.2 | 0.8 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.2 | 0.6 | GO:1902728 | positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.2 | 0.8 | GO:0051582 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.2 | 0.2 | GO:0048865 | stem cell fate commitment(GO:0048865) |
| 0.2 | 1.2 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 3.8 | GO:0031034 | myosin filament assembly(GO:0031034) |
| 0.2 | 0.8 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.2 | 1.7 | GO:0071896 | protein localization to adherens junction(GO:0071896) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.2 | 2.9 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 0.8 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.2 | 0.6 | GO:0060366 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 0.2 | 1.3 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.2 | 1.3 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.2 | 1.1 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.2 | 0.7 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.2 | 0.5 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.2 | 0.9 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.2 | 2.5 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.2 | 0.5 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.2 | 11.5 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.2 | 0.7 | GO:1901993 | pachytene(GO:0000239) meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) |
| 0.2 | 1.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.2 | 0.5 | GO:1902868 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.2 | 0.7 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.2 | 1.2 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.2 | 0.8 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.2 | 1.0 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.2 | 0.5 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.2 | 7.6 | GO:0050891 | multicellular organismal water homeostasis(GO:0050891) |
| 0.2 | 0.8 | GO:0060687 | regulation of branching involved in prostate gland morphogenesis(GO:0060687) |
| 0.2 | 0.6 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.2 | 0.6 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.4 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.1 | 1.0 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 3.4 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.1 | 1.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 2.8 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.1 | 0.4 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.1 | 1.3 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.7 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.1 | 0.8 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.1 | 0.4 | GO:0060450 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) positive regulation of hindgut contraction(GO:0060450) regulation of relaxation of smooth muscle(GO:1901080) positive regulation of relaxation of smooth muscle(GO:1901082) |
| 0.1 | 0.3 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 0.1 | 1.0 | GO:0051036 | regulation of endosome size(GO:0051036) receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 0.5 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.1 | 0.8 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 0.6 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 0.2 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 | 1.0 | GO:2000188 | regulation of cholesterol homeostasis(GO:2000188) |
| 0.1 | 1.3 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 0.8 | GO:0051775 | response to redox state(GO:0051775) |
| 0.1 | 2.0 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.6 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.1 | 2.8 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 1.2 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.7 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.9 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.3 | GO:1901738 | regulation of vitamin A metabolic process(GO:1901738) |
| 0.1 | 1.6 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.7 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.9 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 1.6 | GO:0051608 | histamine transport(GO:0051608) |
| 0.1 | 1.7 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 0.6 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.6 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 0.3 | GO:1904690 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.1 | 0.9 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.5 | GO:0070244 | negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.1 | 2.3 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.1 | 1.0 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 0.9 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.1 | 1.0 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.1 | 2.0 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 1.3 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 | 0.5 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 0.5 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 0.3 | GO:0072276 | cell migration involved in vasculogenesis(GO:0035441) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 13.3 | GO:0009166 | nucleotide catabolic process(GO:0009166) |
| 0.1 | 2.9 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 | 0.4 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 0.7 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.4 | GO:0097117 | guanylate kinase-associated protein clustering(GO:0097117) |
| 0.1 | 0.4 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 1.4 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.1 | 0.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 1.3 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 0.1 | GO:0021644 | vagus nerve morphogenesis(GO:0021644) |
| 0.1 | 0.3 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.1 | 1.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 1.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.9 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.8 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 1.4 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 1.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.2 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.1 | 0.5 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.1 | 0.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 2.3 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.1 | 0.4 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.1 | 0.8 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 0.4 | GO:0015791 | polyol transport(GO:0015791) |
| 0.1 | 1.6 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 1.4 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 | 0.8 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.1 | 0.8 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.8 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 1.0 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.3 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 0.2 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.1 | 0.3 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.1 | 1.3 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.6 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 6.6 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 0.2 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.1 | 1.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.6 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 | 0.6 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.1 | 3.8 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 0.3 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 1.5 | GO:0002828 | regulation of type 2 immune response(GO:0002828) |
| 0.1 | 0.2 | GO:0060022 | hard palate development(GO:0060022) |
| 0.1 | 1.0 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.1 | 3.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.2 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.1 | 0.8 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.1 | 0.6 | GO:0030238 | male sex determination(GO:0030238) |
| 0.1 | 1.0 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.1 | 0.5 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 | 0.5 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 2.4 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 0.2 | GO:0071317 | cellular response to isoquinoline alkaloid(GO:0071317) |
| 0.1 | 0.4 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 0.7 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.8 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 1.4 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.2 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.3 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.2 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.3 | GO:0035865 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.0 | 2.4 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.3 | GO:0072093 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.0 | 0.2 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.1 | GO:0070949 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.0 | 2.0 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.0 | 0.8 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 1.7 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.0 | 0.4 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.5 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 | 0.9 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 0.2 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.0 | 0.3 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.1 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 1.1 | GO:0048566 | embryonic digestive tract development(GO:0048566) |
| 0.0 | 0.0 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.0 | 0.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.3 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.0 | 1.2 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.2 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.3 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 1.3 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.9 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 | 0.6 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.5 | GO:0097152 | mesenchymal cell apoptotic process(GO:0097152) |
| 0.0 | 0.1 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.2 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.0 | 0.5 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.6 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 1.7 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.6 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 1.0 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 1.3 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.7 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 1.5 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 5.2 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 0.6 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.0 | 0.1 | GO:0060278 | regulation of ovulation(GO:0060278) |
| 0.0 | 1.2 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 0.3 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.0 | 0.3 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.7 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 8.7 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 1.2 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 1.3 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 0.6 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.2 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.3 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 1.3 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.2 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.0 | 0.3 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.3 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 8.0 | GO:0043434 | response to peptide hormone(GO:0043434) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.1 | GO:1902608 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.0 | 0.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.0 | 0.3 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 0.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 1.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:1901727 | regulation of histone deacetylase activity(GO:1901725) positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.2 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.0 | 0.3 | GO:0034205 | beta-amyloid formation(GO:0034205) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.6 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.5 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.6 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.0 | GO:0050748 | negative regulation of lipoprotein metabolic process(GO:0050748) |
| 0.0 | 0.4 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.7 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.5 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.7 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.1 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 1.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.6 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.1 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.5 | GO:0002209 | behavioral fear response(GO:0001662) behavioral defense response(GO:0002209) |
| 0.0 | 1.1 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.9 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) cerebral cortex GABAergic interneuron development(GO:0021894) interneuron migration(GO:1904936) |
| 0.0 | 1.0 | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) |
| 0.0 | 0.3 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.6 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.3 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.8 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 1.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.0 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.3 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 7.9 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 1.4 | 4.2 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 1.0 | 10.8 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.6 | 4.9 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.4 | 7.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.4 | 1.3 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.4 | 2.8 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.3 | 1.0 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.3 | 0.9 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.3 | 2.0 | GO:0070695 | FHF complex(GO:0070695) |
| 0.3 | 2.9 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.2 | 0.7 | GO:0097635 | Atg12-Atg5-Atg16 complex(GO:0034274) extrinsic component of autophagosome membrane(GO:0097635) |
| 0.2 | 0.7 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.2 | 1.2 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.2 | 2.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 1.7 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.2 | 0.9 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.2 | 7.0 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.2 | 1.7 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.2 | 5.7 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 3.3 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.2 | 1.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 1.6 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.2 | 4.5 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.2 | 0.7 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.2 | 2.8 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 1.0 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 1.0 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 5.1 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.1 | 2.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.8 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.6 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.1 | 0.6 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.1 | 3.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.2 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.1 | 0.5 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 2.0 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.7 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 1.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 1.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 6.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.7 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.3 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.1 | 1.7 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.5 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 1.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.2 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.1 | 0.6 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 1.0 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.8 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.6 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 0.5 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.6 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 5.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.7 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.2 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.1 | 3.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 2.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.6 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.6 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 1.0 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.4 | GO:1990462 | omegasome(GO:1990462) |
| 0.0 | 0.6 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 1.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 1.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 1.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.3 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 2.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 3.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.7 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 7.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 4.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 6.3 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.2 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.3 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 1.9 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 2.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 1.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 2.7 | GO:0097014 | ciliary plasm(GO:0097014) |
| 0.0 | 1.8 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 3.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 3.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 4.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 1.3 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 1.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.8 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.6 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 1.1 | GO:0030117 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 0.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 1.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 2.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.3 | 25.0 | GO:0004454 | ketohexokinase activity(GO:0004454) |
| 3.8 | 11.5 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 3.4 | 10.3 | GO:0036361 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 3.2 | 9.5 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 2.9 | 8.7 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 2.0 | 7.9 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 1.8 | 11.0 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 1.3 | 8.0 | GO:0032810 | sterol response element binding(GO:0032810) |
| 1.2 | 4.6 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 1.1 | 5.6 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.9 | 6.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.9 | 4.3 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.7 | 2.9 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.7 | 2.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.7 | 4.9 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.7 | 4.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.6 | 5.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.6 | 5.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.5 | 4.4 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.5 | 6.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.5 | 1.6 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.5 | 4.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.5 | 4.5 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.5 | 3.0 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.5 | 1.9 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.5 | 1.4 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.5 | 1.4 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.4 | 1.3 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.4 | 1.8 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.4 | 1.6 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.4 | 0.8 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 0.4 | 1.9 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.4 | 1.9 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.4 | 7.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.3 | 1.0 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 0.3 | 1.0 | GO:0004368 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.3 | 1.3 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.3 | 2.9 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.3 | 1.7 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.3 | 2.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.3 | 1.6 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.3 | 0.8 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.3 | 1.5 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.2 | 0.9 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) ethanol binding(GO:0035276) |
| 0.2 | 1.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.2 | 0.8 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.2 | 1.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 3.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.2 | 2.4 | GO:1990239 | sodium:potassium-exchanging ATPase activity(GO:0005391) steroid hormone binding(GO:1990239) |
| 0.2 | 0.6 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.2 | 0.8 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.2 | 3.5 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.2 | 1.5 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 0.9 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 1.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.2 | 1.0 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.2 | 1.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.2 | 1.0 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.2 | 2.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 2.9 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 0.5 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.2 | 0.6 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.2 | 0.5 | GO:0099530 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.2 | 2.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 0.5 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.2 | 0.8 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 1.0 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 1.5 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.1 | 1.0 | GO:0008158 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.1 | 2.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 21.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.9 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.4 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 1.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 1.5 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.5 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 1.5 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 1.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 4.7 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 0.7 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.1 | 1.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.5 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.1 | 1.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.8 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 0.7 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 5.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 4.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.3 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.1 | 0.4 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 2.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 2.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.5 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.1 | 0.6 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.3 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 2.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.7 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 1.0 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.1 | 0.7 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.3 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 2.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.9 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 7.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 3.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 5.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 0.9 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.8 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.1 | 3.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.3 | GO:0051381 | histamine binding(GO:0051381) |
| 0.1 | 1.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.5 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.9 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.1 | 2.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 2.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 1.0 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 0.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.9 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.8 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 1.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.7 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.7 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 2.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.7 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 0.8 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.2 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.1 | 0.7 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.5 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 0.6 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 2.0 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 1.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.8 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 4.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.6 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 1.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.2 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 2.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 3.1 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.9 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 0.5 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.3 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.5 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.2 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 1.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.9 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.2 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 1.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 2.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.6 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.0 | 1.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 5.3 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.4 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.8 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.5 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.2 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 4.4 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 1.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 1.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.8 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.5 | GO:0042166 | acetylcholine-gated cation channel activity(GO:0022848) acetylcholine binding(GO:0042166) |
| 0.0 | 1.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 15.6 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.1 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.8 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.4 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.5 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 5.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 0.8 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 1.0 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 4.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 6.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 5.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 4.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 2.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 2.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 2.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 3.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 1.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 0.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 0.9 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.6 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.9 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.9 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 3.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 8.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 4.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 2.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.8 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 1.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 5.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 1.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.9 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.8 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.5 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.3 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.3 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 2.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 11.0 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.5 | 5.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 18.4 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.3 | 4.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.3 | 7.3 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.2 | 5.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 8.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 2.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 0.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 1.5 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 0.8 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 4.8 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.1 | 13.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.9 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.9 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 2.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 2.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 12.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 1.7 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 1.6 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 10.3 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 2.5 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.1 | 1.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 3.6 | REACTOME PKB MEDIATED EVENTS | Genes involved in PKB-mediated events |
| 0.1 | 1.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 2.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 21.7 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.1 | 2.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 4.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 1.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 0.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 3.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.1 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 2.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 1.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 0.5 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.5 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 1.0 | REACTOME SIGNALLING BY NGF | Genes involved in Signalling by NGF |
| 0.0 | 1.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.5 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 2.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 2.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.8 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.8 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 1.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.4 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.7 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 2.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.8 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 1.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 1.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.0 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.8 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 2.7 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 1.0 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.7 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 2.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.2 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.2 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |