Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
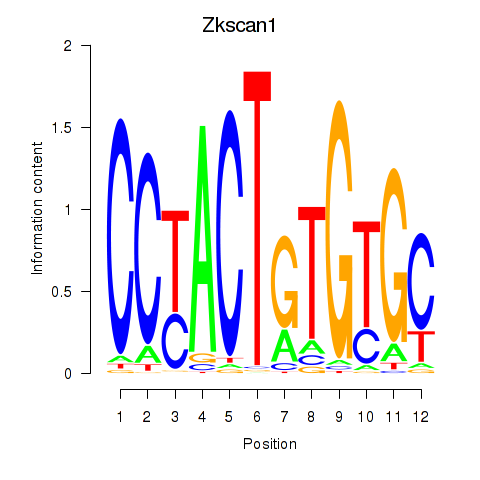
Results for Zkscan1
Z-value: 1.82
Transcription factors associated with Zkscan1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zkscan1
|
ENSMUSG00000029729.13 | Zkscan1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zkscan1 | mm39_v1_chr5_+_138083345_138083410 | -0.32 | 6.1e-02 | Click! |
Activity profile of Zkscan1 motif
Sorted Z-values of Zkscan1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zkscan1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_79716876 | 11.26 |
ENSMUST00000166201.2
|
Prtn3
|
proteinase 3 |
| chr17_-_33937565 | 8.59 |
ENSMUST00000174040.2
ENSMUST00000173015.8 ENSMUST00000066121.13 ENSMUST00000186022.7 ENSMUST00000173329.8 ENSMUST00000172767.9 |
Marchf2
|
membrane associated ring-CH-type finger 2 |
| chr15_-_74983786 | 8.20 |
ENSMUST00000191451.2
ENSMUST00000100542.10 |
Ly6c2
|
lymphocyte antigen 6 complex, locus C2 |
| chr8_-_72178340 | 6.78 |
ENSMUST00000153800.8
ENSMUST00000146100.8 |
Fcho1
|
FCH domain only 1 |
| chr15_-_78456898 | 5.97 |
ENSMUST00000043214.8
|
Rac2
|
Rac family small GTPase 2 |
| chr2_+_84564394 | 5.68 |
ENSMUST00000238573.2
ENSMUST00000090729.9 |
Ypel4
|
yippee like 4 |
| chr17_+_29042640 | 5.48 |
ENSMUST00000233088.2
ENSMUST00000233182.2 ENSMUST00000233520.2 |
Brpf3
|
bromodomain and PHD finger containing, 3 |
| chr9_+_51124983 | 5.31 |
ENSMUST00000034554.9
|
Pou2af1
|
POU domain, class 2, associating factor 1 |
| chrX_-_48886577 | 5.27 |
ENSMUST00000033442.14
ENSMUST00000114891.2 |
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr15_+_75027089 | 5.11 |
ENSMUST00000190262.2
|
Ly6g
|
lymphocyte antigen 6 complex, locus G |
| chr4_-_140805613 | 4.59 |
ENSMUST00000030760.15
|
Necap2
|
NECAP endocytosis associated 2 |
| chr15_-_74920518 | 4.42 |
ENSMUST00000185372.2
ENSMUST00000187347.7 ENSMUST00000188845.7 ENSMUST00000185200.7 ENSMUST00000179762.8 ENSMUST00000191216.7 ENSMUST00000065408.16 |
Ly6c1
|
lymphocyte antigen 6 complex, locus C1 |
| chr17_+_29042544 | 4.38 |
ENSMUST00000140587.9
|
Brpf3
|
bromodomain and PHD finger containing, 3 |
| chr7_-_132415257 | 4.29 |
ENSMUST00000097999.9
|
Fam53b
|
family with sequence similarity 53, member B |
| chr8_+_95712151 | 4.10 |
ENSMUST00000212799.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr19_-_61216834 | 4.03 |
ENSMUST00000076046.7
|
Csf2ra
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr12_-_79054050 | 3.67 |
ENSMUST00000056660.13
ENSMUST00000174721.8 |
Tmem229b
|
transmembrane protein 229B |
| chr9_-_103357564 | 3.42 |
ENSMUST00000124310.5
|
Bfsp2
|
beaded filament structural protein 2, phakinin |
| chr13_+_50571889 | 3.33 |
ENSMUST00000046974.5
|
Fbxw17
|
F-box and WD-40 domain protein 17 |
| chr2_+_118428690 | 3.28 |
ENSMUST00000038341.8
|
Bub1b
|
BUB1B, mitotic checkpoint serine/threonine kinase |
| chr10_-_79624758 | 3.26 |
ENSMUST00000020573.13
|
Prss57
|
protease, serine 57 |
| chr5_+_125518736 | 3.22 |
ENSMUST00000198811.2
|
Bri3bp
|
Bri3 binding protein |
| chr10_-_81360059 | 3.17 |
ENSMUST00000043709.8
|
Gna15
|
guanine nucleotide binding protein, alpha 15 |
| chr6_-_83513222 | 3.12 |
ENSMUST00000075161.12
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr5_-_140307280 | 3.01 |
ENSMUST00000031534.9
|
Mad1l1
|
MAD1 mitotic arrest deficient 1-like 1 |
| chr7_-_132414823 | 3.00 |
ENSMUST00000106165.8
|
Fam53b
|
family with sequence similarity 53, member B |
| chr7_-_126399778 | 2.95 |
ENSMUST00000141355.4
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chrX_+_20736405 | 2.90 |
ENSMUST00000115342.10
ENSMUST00000009530.5 |
Timp1
|
tissue inhibitor of metalloproteinase 1 |
| chr6_+_47854138 | 2.80 |
ENSMUST00000061890.8
|
Zfp282
|
zinc finger protein 282 |
| chr12_+_111387023 | 2.78 |
ENSMUST00000220852.2
|
Exoc3l4
|
exocyst complex component 3-like 4 |
| chr2_+_156681927 | 2.39 |
ENSMUST00000081335.13
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr2_+_156681991 | 2.31 |
ENSMUST00000073352.10
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr6_-_83513184 | 2.30 |
ENSMUST00000205926.2
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr7_-_97811525 | 2.10 |
ENSMUST00000107112.2
|
Capn5
|
calpain 5 |
| chr19_+_53298906 | 2.07 |
ENSMUST00000003870.15
|
Mxi1
|
MAX interactor 1, dimerization protein |
| chr19_+_12775938 | 1.98 |
ENSMUST00000025601.8
|
Lpxn
|
leupaxin |
| chr7_+_96730763 | 1.95 |
ENSMUST00000004622.7
|
Gab2
|
growth factor receptor bound protein 2-associated protein 2 |
| chr11_+_104468107 | 1.83 |
ENSMUST00000106956.10
|
Myl4
|
myosin, light polypeptide 4 |
| chr15_-_74855264 | 1.80 |
ENSMUST00000023250.11
ENSMUST00000166694.2 |
Ly6i
|
lymphocyte antigen 6 complex, locus I |
| chr16_+_90182895 | 1.75 |
ENSMUST00000065856.8
|
Hunk
|
hormonally upregulated Neu-associated kinase |
| chr9_+_118881838 | 1.48 |
ENSMUST00000051386.13
ENSMUST00000074734.13 |
Vill
|
villin-like |
| chr6_-_122317484 | 1.42 |
ENSMUST00000112600.9
|
Phc1
|
polyhomeotic 1 |
| chr10_-_79390911 | 1.39 |
ENSMUST00000170018.2
ENSMUST00000062855.15 ENSMUST00000165778.8 ENSMUST00000165028.8 |
Mier2
|
MIER family member 2 |
| chr17_+_25105617 | 1.35 |
ENSMUST00000117890.8
ENSMUST00000168265.8 ENSMUST00000120943.8 ENSMUST00000068508.13 ENSMUST00000119829.8 |
Spsb3
|
splA/ryanodine receptor domain and SOCS box containing 3 |
| chr5_+_123532819 | 1.25 |
ENSMUST00000111596.8
ENSMUST00000068237.12 |
Mlxip
|
MLX interacting protein |
| chr11_+_69871952 | 1.22 |
ENSMUST00000108593.8
|
Ctdnep1
|
CTD nuclear envelope phosphatase 1 |
| chr2_+_172235702 | 1.16 |
ENSMUST00000099061.9
ENSMUST00000103073.9 |
Cass4
|
Cas scaffolding protein family member 4 |
| chr7_-_132415408 | 1.15 |
ENSMUST00000134946.3
|
Fam53b
|
family with sequence similarity 53, member B |
| chr11_+_78717398 | 1.15 |
ENSMUST00000147875.9
ENSMUST00000141321.2 |
Lyrm9
|
LYR motif containing 9 |
| chr11_-_69872050 | 1.14 |
ENSMUST00000108594.8
|
Elp5
|
elongator acetyltransferase complex subunit 5 |
| chr7_+_28834276 | 1.13 |
ENSMUST00000161522.8
ENSMUST00000204845.3 ENSMUST00000205027.3 ENSMUST00000204194.3 ENSMUST00000203070.3 ENSMUST00000203380.3 |
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chr3_+_89043440 | 1.12 |
ENSMUST00000047111.13
|
Pklr
|
pyruvate kinase liver and red blood cell |
| chr15_+_101308935 | 1.11 |
ENSMUST00000147662.8
|
Krt7
|
keratin 7 |
| chr2_+_172235820 | 1.08 |
ENSMUST00000109136.3
ENSMUST00000228775.2 |
Cass4
|
Cas scaffolding protein family member 4 |
| chr2_+_31135813 | 1.08 |
ENSMUST00000000199.8
|
Ncs1
|
neuronal calcium sensor 1 |
| chr18_+_61238908 | 1.03 |
ENSMUST00000115268.4
|
Csf1r
|
colony stimulating factor 1 receptor |
| chr5_+_64969679 | 1.01 |
ENSMUST00000166409.6
ENSMUST00000197879.2 |
Klf3
|
Kruppel-like factor 3 (basic) |
| chr2_+_90507552 | 1.01 |
ENSMUST00000057481.7
|
Nup160
|
nucleoporin 160 |
| chr12_+_116369017 | 0.99 |
ENSMUST00000084828.5
ENSMUST00000222469.2 ENSMUST00000221114.2 ENSMUST00000221970.2 |
Ncapg2
|
non-SMC condensin II complex, subunit G2 |
| chr1_+_171668173 | 0.92 |
ENSMUST00000136479.8
|
Cd84
|
CD84 antigen |
| chr8_-_70980584 | 0.90 |
ENSMUST00000138260.8
ENSMUST00000117580.8 |
Kxd1
|
KxDL motif containing 1 |
| chr2_+_112092271 | 0.89 |
ENSMUST00000028553.4
|
Nop10
|
NOP10 ribonucleoprotein |
| chr14_-_52553764 | 0.82 |
ENSMUST00000135523.5
|
Sall2
|
spalt like transcription factor 2 |
| chr4_-_63414188 | 0.81 |
ENSMUST00000063650.10
ENSMUST00000102867.8 ENSMUST00000107393.8 ENSMUST00000084510.8 ENSMUST00000095038.8 ENSMUST00000119294.8 ENSMUST00000095037.2 ENSMUST00000063672.10 |
Whrn
|
whirlin |
| chr5_+_125518609 | 0.80 |
ENSMUST00000049040.14
|
Bri3bp
|
Bri3 binding protein |
| chr19_+_41818409 | 0.75 |
ENSMUST00000087155.5
|
Frat1
|
frequently rearranged in advanced T cell lymphomas |
| chr8_+_4303067 | 0.68 |
ENSMUST00000011981.5
ENSMUST00000208316.2 |
Snapc2
|
small nuclear RNA activating complex, polypeptide 2 |
| chr11_+_23234644 | 0.66 |
ENSMUST00000150750.3
|
Xpo1
|
exportin 1 |
| chr2_+_118730823 | 0.63 |
ENSMUST00000151162.2
|
Bahd1
|
bromo adjacent homology domain containing 1 |
| chrX_+_163156359 | 0.63 |
ENSMUST00000033751.8
|
Vegfd
|
vascular endothelial growth factor D |
| chr4_+_141171965 | 0.62 |
ENSMUST00000006377.13
|
Zbtb17
|
zinc finger and BTB domain containing 17 |
| chr10_+_79889322 | 0.59 |
ENSMUST00000105372.9
|
Gpx4
|
glutathione peroxidase 4 |
| chr13_-_56283331 | 0.58 |
ENSMUST00000045788.9
ENSMUST00000016081.13 |
Macroh2a1
|
macroH2A.1 histone |
| chr19_+_44321531 | 0.57 |
ENSMUST00000058856.9
|
Scd4
|
stearoyl-coenzyme A desaturase 4 |
| chr6_-_122317457 | 0.56 |
ENSMUST00000160843.8
|
Phc1
|
polyhomeotic 1 |
| chr11_+_53992054 | 0.55 |
ENSMUST00000135653.8
|
P4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha II polypeptide |
| chrX_+_158155171 | 0.49 |
ENSMUST00000087143.7
|
Eif1ax
|
eukaryotic translation initiation factor 1A, X-linked |
| chr5_-_134668152 | 0.48 |
ENSMUST00000036125.10
ENSMUST00000202622.4 |
Eif4h
|
eukaryotic translation initiation factor 4H |
| chr11_-_78056347 | 0.48 |
ENSMUST00000017530.4
|
Traf4
|
TNF receptor associated factor 4 |
| chr4_+_132948121 | 0.46 |
ENSMUST00000105910.2
|
Cd164l2
|
CD164 sialomucin-like 2 |
| chr1_-_93563406 | 0.45 |
ENSMUST00000027498.14
|
Stk25
|
serine/threonine kinase 25 (yeast) |
| chr19_+_10914517 | 0.45 |
ENSMUST00000037261.4
|
Ptgdr2
|
prostaglandin D2 receptor 2 |
| chr4_+_101276474 | 0.44 |
ENSMUST00000102780.8
ENSMUST00000106946.8 ENSMUST00000106945.8 |
Ak4
|
adenylate kinase 4 |
| chr1_-_93563201 | 0.40 |
ENSMUST00000133769.8
|
Stk25
|
serine/threonine kinase 25 (yeast) |
| chr11_-_78402931 | 0.37 |
ENSMUST00000052566.8
|
Tmem199
|
transmembrane protein 199 |
| chr15_-_74869684 | 0.35 |
ENSMUST00000190188.2
ENSMUST00000189068.7 ENSMUST00000186526.7 ENSMUST00000187171.2 ENSMUST00000187994.7 |
Ly6a
|
lymphocyte antigen 6 complex, locus A |
| chr7_-_118091135 | 0.35 |
ENSMUST00000178344.3
|
Itpripl2
|
inositol 1,4,5-triphosphate receptor interacting protein-like 2 |
| chr5_-_131567526 | 0.32 |
ENSMUST00000161374.8
|
Auts2
|
autism susceptibility candidate 2 |
| chr9_+_118881926 | 0.31 |
ENSMUST00000131647.2
|
Vill
|
villin-like |
| chr10_-_38998272 | 0.31 |
ENSMUST00000136546.8
|
Fam229b
|
family with sequence similarity 229, member B |
| chr6_+_57234937 | 0.30 |
ENSMUST00000228297.2
|
Vmn1r15
|
vomeronasal 1 receptor 15 |
| chr14_-_40728839 | 0.28 |
ENSMUST00000152837.2
ENSMUST00000134715.8 |
Prxl2a
|
peroxiredoxin like 2A |
| chr7_-_28661648 | 0.28 |
ENSMUST00000127210.8
|
Actn4
|
actinin alpha 4 |
| chr15_-_82505132 | 0.26 |
ENSMUST00000109515.3
|
Cyp2d34
|
cytochrome P450, family 2, subfamily d, polypeptide 34 |
| chr15_-_79048674 | 0.26 |
ENSMUST00000230261.2
ENSMUST00000040019.5 |
Sox10
|
SRY (sex determining region Y)-box 10 |
| chr18_-_52662917 | 0.26 |
ENSMUST00000171470.8
|
Lox
|
lysyl oxidase |
| chr10_+_90665270 | 0.25 |
ENSMUST00000182202.8
ENSMUST00000182966.8 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr17_+_34258411 | 0.24 |
ENSMUST00000087497.11
ENSMUST00000131134.9 ENSMUST00000235819.2 ENSMUST00000114255.9 ENSMUST00000114252.9 ENSMUST00000237989.2 |
Col11a2
|
collagen, type XI, alpha 2 |
| chr9_+_98305014 | 0.24 |
ENSMUST00000052068.11
|
Rbp1
|
retinol binding protein 1, cellular |
| chr7_-_28661751 | 0.23 |
ENSMUST00000068045.14
ENSMUST00000217157.2 |
Actn4
|
actinin alpha 4 |
| chr10_+_90665639 | 0.22 |
ENSMUST00000179337.9
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr4_+_109263820 | 0.22 |
ENSMUST00000124209.8
|
Ttc39a
|
tetratricopeptide repeat domain 39A |
| chr2_-_91795910 | 0.20 |
ENSMUST00000239257.2
|
Dgkz
|
diacylglycerol kinase zeta |
| chr2_-_127571836 | 0.19 |
ENSMUST00000028856.3
|
Mall
|
mal, T cell differentiation protein-like |
| chr19_-_53932581 | 0.17 |
ENSMUST00000236885.2
ENSMUST00000236098.2 ENSMUST00000236370.2 |
Bbip1
|
BBSome interacting protein 1 |
| chr4_-_97472844 | 0.17 |
ENSMUST00000107067.8
ENSMUST00000107068.9 |
E130114P18Rik
|
RIKEN cDNA E130114P18 gene |
| chr15_+_16778187 | 0.17 |
ENSMUST00000026432.8
|
Cdh9
|
cadherin 9 |
| chr4_+_103476777 | 0.16 |
ENSMUST00000106827.8
|
Dab1
|
disabled 1 |
| chr6_+_83055321 | 0.16 |
ENSMUST00000165164.9
ENSMUST00000092614.9 |
Pcgf1
|
polycomb group ring finger 1 |
| chr9_+_106245792 | 0.16 |
ENSMUST00000172306.3
|
Dusp7
|
dual specificity phosphatase 7 |
| chr1_+_90531183 | 0.16 |
ENSMUST00000186750.2
|
Cops8
|
COP9 signalosome subunit 8 |
| chr15_-_34356567 | 0.15 |
ENSMUST00000179647.2
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr10_+_90665399 | 0.15 |
ENSMUST00000179694.9
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr11_+_120348919 | 0.14 |
ENSMUST00000058370.14
ENSMUST00000175970.8 ENSMUST00000176120.2 |
Ccdc137
|
coiled-coil domain containing 137 |
| chrX_-_103244703 | 0.13 |
ENSMUST00000087879.11
|
Nexmif
|
neurite extension and migration factor |
| chr2_-_73410632 | 0.12 |
ENSMUST00000028515.4
|
Chrna1
|
cholinergic receptor, nicotinic, alpha polypeptide 1 (muscle) |
| chr19_-_53932867 | 0.12 |
ENSMUST00000235688.2
ENSMUST00000235348.2 |
Bbip1
|
BBSome interacting protein 1 |
| chr11_-_78313043 | 0.11 |
ENSMUST00000001122.6
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr17_+_5045178 | 0.11 |
ENSMUST00000092723.11
ENSMUST00000232180.2 ENSMUST00000115797.9 |
Arid1b
|
AT rich interactive domain 1B (SWI-like) |
| chr16_-_10265204 | 0.10 |
ENSMUST00000051118.7
|
Tvp23a
|
trans-golgi network vesicle protein 23A |
| chr8_-_4375320 | 0.10 |
ENSMUST00000098950.6
|
Elavl1
|
ELAV (embryonic lethal, abnormal vision)-like 1 (Hu antigen R) |
| chr4_+_73931679 | 0.09 |
ENSMUST00000098006.9
ENSMUST00000084474.6 |
Frmd3
|
FERM domain containing 3 |
| chr19_+_41471395 | 0.09 |
ENSMUST00000237208.2
ENSMUST00000238398.2 |
Lcor
|
ligand dependent nuclear receptor corepressor |
| chr12_+_112586501 | 0.08 |
ENSMUST00000180015.9
|
Adssl1
|
adenylosuccinate synthetase like 1 |
| chr2_+_153742294 | 0.07 |
ENSMUST00000088955.12
ENSMUST00000135501.3 |
Bpifb6
|
BPI fold containing family B, member 6 |
| chr6_+_90246088 | 0.07 |
ENSMUST00000058039.3
|
Vmn1r54
|
vomeronasal 1 receptor 54 |
| chr19_+_43678109 | 0.07 |
ENSMUST00000081079.6
|
Entpd7
|
ectonucleoside triphosphate diphosphohydrolase 7 |
| chr6_+_134012602 | 0.06 |
ENSMUST00000081028.13
ENSMUST00000111963.8 |
Etv6
|
ets variant 6 |
| chr4_+_126156118 | 0.06 |
ENSMUST00000030660.9
|
Trappc3
|
trafficking protein particle complex 3 |
| chr11_+_42310557 | 0.05 |
ENSMUST00000007797.10
|
Gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 2 |
| chr14_+_34395845 | 0.04 |
ENSMUST00000048263.14
|
Wapl
|
WAPL cohesin release factor |
| chr19_+_5138562 | 0.04 |
ENSMUST00000238093.2
ENSMUST00000025811.6 ENSMUST00000237025.2 |
Yif1a
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chrX_-_103244784 | 0.03 |
ENSMUST00000118314.8
|
Nexmif
|
neurite extension and migration factor |
| chr17_+_47604995 | 0.03 |
ENSMUST00000190020.4
|
Trerf1
|
transcriptional regulating factor 1 |
| chr10_+_94350687 | 0.02 |
ENSMUST00000065060.12
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 11.3 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.7 | 6.6 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.7 | 3.3 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.4 | 3.0 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.3 | 2.9 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.3 | 2.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.2 | 0.6 | GO:0032685 | negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) |
| 0.2 | 3.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.2 | 6.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.2 | 4.7 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.2 | 0.9 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.2 | 1.0 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.2 | 2.9 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 4.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.2 | 2.8 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 8.1 | GO:0035411 | catenin import into nucleus(GO:0035411) |
| 0.2 | 3.2 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.2 | 2.0 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.1 | 0.8 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 0.5 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.1 | 1.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 9.9 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 0.6 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.3 | GO:0098582 | innate vocalization behavior(GO:0098582) positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.1 | 0.2 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.1 | 0.2 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 2.0 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 0.4 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 0.8 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.1 | 0.7 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.6 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 0.6 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 1.0 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.2 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 | 0.1 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.0 | 0.4 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 1.8 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.6 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.3 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.5 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 2.1 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.6 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 1.0 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 1.2 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.8 | GO:0000578 | embryonic axis specification(GO:0000578) |
| 0.0 | 0.3 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.0 | 1.4 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.9 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.7 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 9.9 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 1.1 | 3.3 | GO:0034774 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.4 | 1.2 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.4 | 4.1 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.3 | 1.0 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.2 | 3.3 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.2 | 0.9 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.2 | 2.0 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.2 | 1.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.2 | 0.8 | GO:0002142 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.1 | 2.9 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 3.8 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.1 | 2.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.6 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 19.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.0 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 6.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 1.0 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.6 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 1.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.7 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 2.0 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 3.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 4.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 2.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 5.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.8 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 6.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.8 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.3 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 4.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 3.8 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 10.4 | GO:0031012 | extracellular matrix(GO:0031012) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.6 | 5.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.3 | 1.0 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.3 | 6.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.3 | 2.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 1.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.4 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.1 | 0.4 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 3.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 2.0 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.9 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 2.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.6 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 0.6 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 5.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 2.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.5 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.1 | 0.6 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.1 | 2.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 4.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.7 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 9.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 1.0 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 1.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.6 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 1.4 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 1.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 4.1 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.0 | 2.6 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 1.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.0 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.8 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 15.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 4.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 5.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 4.9 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 3.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 0.7 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 5.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 2.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.1 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 5.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 3.3 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.1 | 16.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 4.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 6.0 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 1.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 0.8 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 1.8 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 3.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.7 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 1.0 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |