Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Zscan4c
Z-value: 0.75
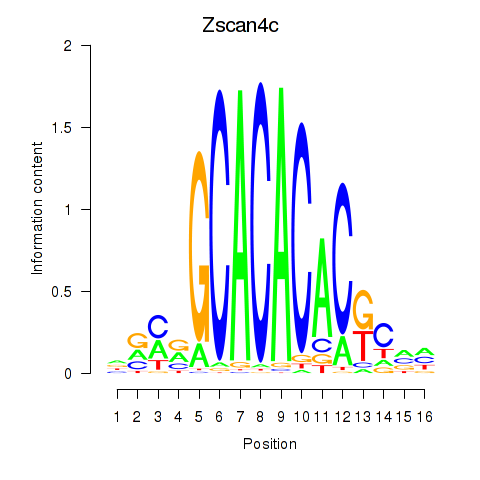
Transcription factors associated with Zscan4c
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zscan4c
|
ENSMUSG00000054272.7 | Zscan4c |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zscan4c | mm39_v1_chr7_+_10739672_10739672 | 0.08 | 6.6e-01 | Click! |
Activity profile of Zscan4c motif
Sorted Z-values of Zscan4c motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zscan4c
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_28779678 | 2.81 |
ENSMUST00000114785.3
ENSMUST00000025062.5 |
Clps
|
colipase, pancreatic |
| chr4_-_46991842 | 2.22 |
ENSMUST00000107749.4
|
Gabbr2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr2_+_102489558 | 2.18 |
ENSMUST00000111213.8
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr4_+_137589548 | 2.17 |
ENSMUST00000102518.10
|
Ece1
|
endothelin converting enzyme 1 |
| chr2_+_102488985 | 2.06 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr10_+_87694117 | 2.01 |
ENSMUST00000122386.8
|
Igf1
|
insulin-like growth factor 1 |
| chr5_+_65127412 | 1.39 |
ENSMUST00000031080.15
|
Fam114a1
|
family with sequence similarity 114, member A1 |
| chr9_-_44710480 | 1.37 |
ENSMUST00000214833.2
ENSMUST00000213972.2 ENSMUST00000214431.2 ENSMUST00000213363.2 ENSMUST00000114705.9 ENSMUST00000002100.8 |
Tmem25
|
transmembrane protein 25 |
| chr2_+_57128309 | 1.32 |
ENSMUST00000112618.9
ENSMUST00000028167.3 |
Gpd2
|
glycerol phosphate dehydrogenase 2, mitochondrial |
| chr2_-_165997551 | 1.31 |
ENSMUST00000109249.9
ENSMUST00000146497.9 |
Sulf2
|
sulfatase 2 |
| chr6_+_91661074 | 1.22 |
ENSMUST00000205480.2
ENSMUST00000206545.2 |
Slc6a6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
| chr9_+_37466989 | 1.20 |
ENSMUST00000213126.2
|
Siae
|
sialic acid acetylesterase |
| chr16_-_38533597 | 1.11 |
ENSMUST00000023487.5
|
Arhgap31
|
Rho GTPase activating protein 31 |
| chr6_+_124470053 | 1.08 |
ENSMUST00000049124.10
|
C1rl
|
complement component 1, r subcomponent-like |
| chr15_+_44482545 | 0.98 |
ENSMUST00000227691.2
|
Ebag9
|
estrogen receptor-binding fragment-associated gene 9 |
| chr7_-_30754792 | 0.97 |
ENSMUST00000206328.2
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr15_+_44482667 | 0.96 |
ENSMUST00000228648.2
ENSMUST00000226165.2 |
Ebag9
|
estrogen receptor-binding fragment-associated gene 9 |
| chr2_-_165997179 | 0.94 |
ENSMUST00000088086.4
|
Sulf2
|
sulfatase 2 |
| chr1_+_182591425 | 0.93 |
ENSMUST00000155229.7
ENSMUST00000153348.8 |
Susd4
|
sushi domain containing 4 |
| chr19_-_47452557 | 0.87 |
ENSMUST00000111800.4
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr2_+_30254239 | 0.86 |
ENSMUST00000077977.14
ENSMUST00000140075.9 ENSMUST00000142801.8 ENSMUST00000100214.10 |
Miga2
|
mitoguardin 2 |
| chr2_+_169474916 | 0.85 |
ENSMUST00000109159.3
|
Tshz2
|
teashirt zinc finger family member 2 |
| chr5_-_96309600 | 0.84 |
ENSMUST00000129646.8
ENSMUST00000113005.9 ENSMUST00000154500.2 ENSMUST00000141383.8 |
Cnot6l
|
CCR4-NOT transcription complex, subunit 6-like |
| chr13_-_9815350 | 0.82 |
ENSMUST00000110636.9
ENSMUST00000152725.8 |
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr6_+_91661034 | 0.76 |
ENSMUST00000032185.9
|
Slc6a6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
| chr6_+_48570817 | 0.74 |
ENSMUST00000154010.8
ENSMUST00000009420.15 ENSMUST00000163452.7 ENSMUST00000118229.2 ENSMUST00000135151.3 |
Repin1
|
replication initiator 1 |
| chr1_+_87983189 | 0.74 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr1_+_87983099 | 0.74 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr15_+_44482944 | 0.72 |
ENSMUST00000022964.9
|
Ebag9
|
estrogen receptor-binding fragment-associated gene 9 |
| chr2_+_169475436 | 0.68 |
ENSMUST00000109157.2
|
Tshz2
|
teashirt zinc finger family member 2 |
| chr13_-_9814901 | 0.67 |
ENSMUST00000223421.2
ENSMUST00000128658.8 |
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr13_-_9815173 | 0.67 |
ENSMUST00000062658.15
ENSMUST00000222358.2 |
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr12_+_52551092 | 0.66 |
ENSMUST00000217820.2
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr15_-_37458768 | 0.65 |
ENSMUST00000116445.9
|
Ncald
|
neurocalcin delta |
| chr18_+_64473091 | 0.58 |
ENSMUST00000175965.10
|
Onecut2
|
one cut domain, family member 2 |
| chr17_-_63188139 | 0.56 |
ENSMUST00000078839.5
ENSMUST00000076840.12 |
Efna5
|
ephrin A5 |
| chr16_+_44914397 | 0.56 |
ENSMUST00000061050.6
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr5_+_102629365 | 0.52 |
ENSMUST00000112854.8
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr19_-_21449916 | 0.50 |
ENSMUST00000087600.10
|
Gda
|
guanine deaminase |
| chr13_+_58955675 | 0.50 |
ENSMUST00000224402.2
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr13_-_9814979 | 0.50 |
ENSMUST00000110634.8
|
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr16_-_94091101 | 0.49 |
ENSMUST00000227141.2
|
Hlcs
|
holocarboxylase synthetase (biotin- [propriony-Coenzyme A-carboxylase (ATP-hydrolysing)] ligase) |
| chr9_-_50639367 | 0.48 |
ENSMUST00000117646.8
|
Dixdc1
|
DIX domain containing 1 |
| chr12_+_52550775 | 0.48 |
ENSMUST00000219443.2
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr19_-_43879031 | 0.48 |
ENSMUST00000212048.2
|
Dnmbp
|
dynamin binding protein |
| chr14_+_65596070 | 0.47 |
ENSMUST00000066994.7
|
Zfp395
|
zinc finger protein 395 |
| chr2_+_179684288 | 0.45 |
ENSMUST00000041126.9
|
Ss18l1
|
SS18, nBAF chromatin remodeling complex subunit like 1 |
| chr13_-_9815276 | 0.44 |
ENSMUST00000130151.8
|
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr5_-_134776101 | 0.44 |
ENSMUST00000015138.13
|
Eln
|
elastin |
| chr10_+_18345706 | 0.40 |
ENSMUST00000162891.8
ENSMUST00000100054.4 |
Nhsl1
|
NHS-like 1 |
| chr3_-_51184895 | 0.40 |
ENSMUST00000108051.8
ENSMUST00000108053.9 |
Elf2
|
E74-like factor 2 |
| chr4_+_118384426 | 0.39 |
ENSMUST00000030261.6
|
2610528J11Rik
|
RIKEN cDNA 2610528J11 gene |
| chr7_+_112278520 | 0.37 |
ENSMUST00000084705.13
ENSMUST00000239442.2 ENSMUST00000239404.2 ENSMUST00000059768.18 |
Tead1
|
TEA domain family member 1 |
| chr19_+_8641369 | 0.37 |
ENSMUST00000035444.10
ENSMUST00000163785.2 |
Chrm1
|
cholinergic receptor, muscarinic 1, CNS |
| chr2_+_68691778 | 0.35 |
ENSMUST00000028426.9
|
Cers6
|
ceramide synthase 6 |
| chr1_+_182591771 | 0.34 |
ENSMUST00000193660.6
|
Susd4
|
sushi domain containing 4 |
| chr2_-_35995283 | 0.32 |
ENSMUST00000112960.8
ENSMUST00000112967.12 ENSMUST00000112963.8 |
Lhx6
|
LIM homeobox protein 6 |
| chr17_+_75772475 | 0.31 |
ENSMUST00000095204.6
|
Rasgrp3
|
RAS, guanyl releasing protein 3 |
| chr19_-_38807600 | 0.29 |
ENSMUST00000025963.8
|
Noc3l
|
NOC3 like DNA replication regulator |
| chr9_+_43978369 | 0.29 |
ENSMUST00000177054.8
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr15_-_77037756 | 0.27 |
ENSMUST00000227314.2
ENSMUST00000227930.2 ENSMUST00000227533.2 |
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr15_-_77037972 | 0.27 |
ENSMUST00000111581.4
ENSMUST00000166610.8 |
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr5_-_44956932 | 0.26 |
ENSMUST00000199261.2
ENSMUST00000199534.5 |
Ldb2
|
LIM domain binding 2 |
| chr9_-_50650663 | 0.26 |
ENSMUST00000117093.2
ENSMUST00000121634.8 |
Dixdc1
|
DIX domain containing 1 |
| chr18_-_40352372 | 0.25 |
ENSMUST00000025364.6
|
Yipf5
|
Yip1 domain family, member 5 |
| chr7_+_141276575 | 0.25 |
ENSMUST00000185406.8
|
Muc2
|
mucin 2 |
| chr3_-_51184730 | 0.24 |
ENSMUST00000195432.2
ENSMUST00000091144.11 ENSMUST00000156983.3 |
Elf2
|
E74-like factor 2 |
| chr15_-_77037926 | 0.22 |
ENSMUST00000228087.2
|
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr19_-_57227742 | 0.22 |
ENSMUST00000111559.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr5_-_44956981 | 0.21 |
ENSMUST00000070748.10
|
Ldb2
|
LIM domain binding 2 |
| chr5_-_44957016 | 0.19 |
ENSMUST00000199256.5
|
Ldb2
|
LIM domain binding 2 |
| chr19_+_28941292 | 0.19 |
ENSMUST00000045674.4
|
Plpp6
|
phospholipid phosphatase 6 |
| chr16_-_9812410 | 0.18 |
ENSMUST00000115835.8
|
Grin2a
|
glutamate receptor, ionotropic, NMDA2A (epsilon 1) |
| chr9_+_21279802 | 0.16 |
ENSMUST00000214474.2
|
Ilf3
|
interleukin enhancer binding factor 3 |
| chr18_+_37108994 | 0.16 |
ENSMUST00000193984.2
|
Gm37388
|
predicted gene, 37388 |
| chr2_-_115895528 | 0.15 |
ENSMUST00000028639.13
ENSMUST00000102538.11 |
Meis2
|
Meis homeobox 2 |
| chr2_-_35994819 | 0.13 |
ENSMUST00000148852.4
|
Lhx6
|
LIM homeobox protein 6 |
| chr13_+_58955506 | 0.13 |
ENSMUST00000079828.7
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr16_-_78373510 | 0.12 |
ENSMUST00000231973.2
ENSMUST00000232528.2 ENSMUST00000114220.9 |
D16Ertd472e
|
DNA segment, Chr 16, ERATO Doi 472, expressed |
| chr5_-_129030367 | 0.12 |
ENSMUST00000111346.6
ENSMUST00000200470.5 |
Rimbp2
|
RIMS binding protein 2 |
| chr2_+_68691902 | 0.12 |
ENSMUST00000176018.2
|
Cers6
|
ceramide synthase 6 |
| chr16_-_78373537 | 0.11 |
ENSMUST00000232052.2
ENSMUST00000114219.8 ENSMUST00000114218.2 |
D16Ertd472e
|
DNA segment, Chr 16, ERATO Doi 472, expressed |
| chr10_+_127478844 | 0.10 |
ENSMUST00000092074.12
ENSMUST00000120279.2 |
Stat6
|
signal transducer and activator of transcription 6 |
| chr8_-_49008881 | 0.09 |
ENSMUST00000110345.8
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr9_+_89791943 | 0.08 |
ENSMUST00000189545.2
ENSMUST00000034909.11 ENSMUST00000034912.6 |
Rasgrf1
|
RAS protein-specific guanine nucleotide-releasing factor 1 |
| chr7_-_98010478 | 0.07 |
ENSMUST00000094161.11
ENSMUST00000164726.8 ENSMUST00000206414.2 ENSMUST00000167405.3 |
Tsku
|
tsukushi, small leucine rich proteoglycan |
| chr9_-_87613301 | 0.07 |
ENSMUST00000034991.8
|
Tbx18
|
T-box18 |
| chr11_-_113641980 | 0.07 |
ENSMUST00000153453.2
|
Cdc42ep4
|
CDC42 effector protein (Rho GTPase binding) 4 |
| chrX_-_160942713 | 0.06 |
ENSMUST00000087085.10
|
Nhs
|
NHS actin remodeling regulator |
| chr9_+_36744016 | 0.06 |
ENSMUST00000214772.2
|
Fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr10_-_108535970 | 0.05 |
ENSMUST00000218023.2
|
Gm5136
|
predicted gene 5136 |
| chr7_+_144450260 | 0.05 |
ENSMUST00000033389.7
ENSMUST00000207229.2 |
Fgf15
|
fibroblast growth factor 15 |
| chr6_-_122317156 | 0.05 |
ENSMUST00000159384.8
|
Phc1
|
polyhomeotic 1 |
| chr9_-_31824758 | 0.04 |
ENSMUST00000116615.5
|
Barx2
|
BarH-like homeobox 2 |
| chr18_-_79152504 | 0.04 |
ENSMUST00000025430.11
|
Setbp1
|
SET binding protein 1 |
| chr15_-_85462271 | 0.03 |
ENSMUST00000229191.2
|
Wnt7b
|
wingless-type MMTV integration site family, member 7B |
| chr7_-_98010534 | 0.02 |
ENSMUST00000165257.8
|
Tsku
|
tsukushi, small leucine rich proteoglycan |
| chr8_+_63404395 | 0.02 |
ENSMUST00000119068.8
|
Spock3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 3 |
| chr5_-_129030111 | 0.02 |
ENSMUST00000196085.5
|
Rimbp2
|
RIMS binding protein 2 |
| chr10_+_81128795 | 0.02 |
ENSMUST00000163075.8
ENSMUST00000105327.10 ENSMUST00000045469.15 |
Pip5k1c
|
phosphatidylinositol-4-phosphate 5-kinase, type 1 gamma |
| chr7_+_139827152 | 0.02 |
ENSMUST00000164583.8
ENSMUST00000093984.3 |
Scart2
|
scavenger receptor family member expressed on T cells 2 |
| chr15_-_98705791 | 0.01 |
ENSMUST00000075444.8
|
Ddn
|
dendrin |
| chr14_+_53257873 | 0.00 |
ENSMUST00000196756.2
|
Trav7d-6
|
T cell receptor alpha variable 7D-6 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.7 | 2.0 | GO:0001762 | beta-alanine transport(GO:0001762) taurine transport(GO:0015734) |
| 0.6 | 4.2 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.4 | 2.3 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.3 | 1.3 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 0.3 | 2.0 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.1 | 1.3 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 0.5 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.1 | 1.5 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.5 | GO:0071110 | protein biotinylation(GO:0009305) response to biotin(GO:0070781) histone biotinylation(GO:0071110) |
| 0.1 | 0.6 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.1 | 1.0 | GO:1903278 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.1 | 0.4 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.8 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 0.9 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.1 | 0.5 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 3.1 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.1 | 0.8 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 2.8 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 0.7 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.5 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 2.2 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.6 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.4 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.6 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.1 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.9 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.2 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.0 | 0.7 | GO:2001273 | regulation of glucose import in response to insulin stimulus(GO:2001273) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.4 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 1.1 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 | 0.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.3 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.0 | GO:0072054 | renal outer medulla development(GO:0072054) |
| 0.0 | 0.0 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.3 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.3 | 1.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 2.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.2 | 2.0 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.4 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 0.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 4.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.8 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.6 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.4 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.5 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.9 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.7 | 2.0 | GO:0005369 | beta-alanine transmembrane transporter activity(GO:0001761) taurine transmembrane transporter activity(GO:0005368) taurine:sodium symporter activity(GO:0005369) |
| 0.4 | 1.3 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.4 | 2.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.3 | 2.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 0.6 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.5 | GO:0004077 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.1 | 0.6 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.1 | 0.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.9 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 2.0 | GO:0043539 | insulin-like growth factor receptor binding(GO:0005159) protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 2.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.7 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 1.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 1.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 1.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 2.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.6 | PID SHP2 PATHWAY | SHP2 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 2.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.8 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 4.2 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |