Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
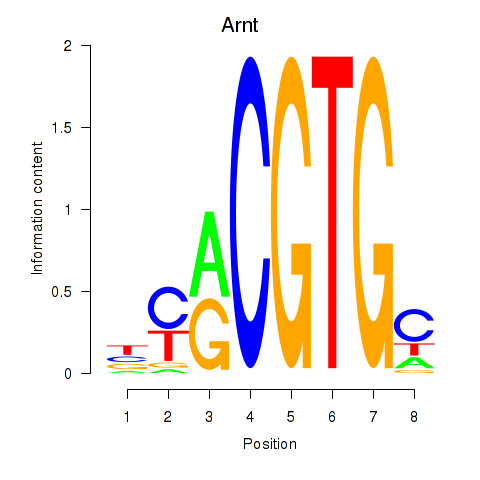
Results for Arnt
Z-value: 1.01
Transcription factors associated with Arnt
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Arnt
|
ENSMUSG00000015522.19 | aryl hydrocarbon receptor nuclear translocator |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Arnt | mm39_v1_chr3_+_95341698_95341739 | -0.54 | 6.0e-04 | Click! |
Activity profile of Arnt motif
Sorted Z-values of Arnt motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_13209141 | 5.92 |
ENSMUST00000033824.8
|
Lamp1
|
lysosomal-associated membrane protein 1 |
| chr1_-_121255448 | 4.94 |
ENSMUST00000186915.2
ENSMUST00000160968.8 ENSMUST00000162582.2 |
Insig2
|
insulin induced gene 2 |
| chr1_-_121255753 | 4.74 |
ENSMUST00000003818.14
|
Insig2
|
insulin induced gene 2 |
| chr1_-_121255400 | 4.62 |
ENSMUST00000159085.8
ENSMUST00000159125.2 ENSMUST00000161818.2 |
Insig2
|
insulin induced gene 2 |
| chr19_+_7034149 | 3.95 |
ENSMUST00000040261.7
|
Macrod1
|
mono-ADP ribosylhydrolase 1 |
| chr1_-_121255503 | 3.85 |
ENSMUST00000160688.2
|
Insig2
|
insulin induced gene 2 |
| chr3_-_95789505 | 3.57 |
ENSMUST00000159863.2
ENSMUST00000159739.8 ENSMUST00000036418.10 ENSMUST00000161866.8 |
Ciart
|
circadian associated repressor of transcription |
| chr8_+_105996469 | 3.50 |
ENSMUST00000172525.8
ENSMUST00000174837.8 ENSMUST00000173859.2 |
Hsf4
|
heat shock transcription factor 4 |
| chr1_+_166081755 | 3.48 |
ENSMUST00000194964.6
ENSMUST00000192638.6 ENSMUST00000192426.6 ENSMUST00000195557.6 ENSMUST00000192732.6 ENSMUST00000193860.2 |
Ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr11_-_98666159 | 3.17 |
ENSMUST00000064941.7
|
Nr1d1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr8_+_105996419 | 3.15 |
ENSMUST00000036127.9
ENSMUST00000163734.9 |
Hsf4
|
heat shock transcription factor 4 |
| chr5_-_65593217 | 2.98 |
ENSMUST00000031103.14
|
Ugdh
|
UDP-glucose dehydrogenase |
| chr17_+_37253802 | 2.88 |
ENSMUST00000040498.12
|
Rnf39
|
ring finger protein 39 |
| chr1_+_166081664 | 2.80 |
ENSMUST00000111416.7
|
Ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr11_-_116089866 | 2.60 |
ENSMUST00000066587.12
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr16_-_97763780 | 2.48 |
ENSMUST00000232187.2
ENSMUST00000231263.2 ENSMUST00000052089.9 ENSMUST00000063605.15 ENSMUST00000113734.9 ENSMUST00000231560.2 ENSMUST00000232165.2 |
Zbtb21
C2cd2
|
zinc finger and BTB domain containing 21 C2 calcium-dependent domain containing 2 |
| chr7_-_16348862 | 2.42 |
ENSMUST00000171937.2
ENSMUST00000075845.11 |
Arhgap35
|
Rho GTPase activating protein 35 |
| chr7_+_28466160 | 2.36 |
ENSMUST00000122915.8
ENSMUST00000072965.5 ENSMUST00000170068.9 |
Sirt2
|
sirtuin 2 |
| chr3_+_89960121 | 2.31 |
ENSMUST00000160640.8
ENSMUST00000029552.13 ENSMUST00000162114.8 ENSMUST00000068798.13 |
4933434E20Rik
|
RIKEN cDNA 4933434E20 gene |
| chr3_-_95789338 | 2.25 |
ENSMUST00000161994.2
|
Ciart
|
circadian associated repressor of transcription |
| chr16_+_44979086 | 2.23 |
ENSMUST00000023343.4
|
Atg3
|
autophagy related 3 |
| chr7_+_46445512 | 2.14 |
ENSMUST00000006774.11
ENSMUST00000165031.8 |
Gtf2h1
|
general transcription factor II H, polypeptide 1 |
| chr14_-_78970160 | 2.09 |
ENSMUST00000226342.3
|
Dgkh
|
diacylglycerol kinase, eta |
| chr10_-_62322551 | 2.08 |
ENSMUST00000105447.11
|
Vps26a
|
VPS26 retromer complex component A |
| chr7_-_28465870 | 2.02 |
ENSMUST00000085851.12
ENSMUST00000032815.11 |
Nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, beta |
| chr11_+_102652228 | 1.98 |
ENSMUST00000103081.10
ENSMUST00000068150.7 |
Adam11
|
a disintegrin and metallopeptidase domain 11 |
| chr5_+_45650821 | 1.90 |
ENSMUST00000198534.2
|
Lap3
|
leucine aminopeptidase 3 |
| chr14_+_4230569 | 1.87 |
ENSMUST00000090543.6
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr16_-_4238280 | 1.81 |
ENSMUST00000120080.8
|
Adcy9
|
adenylate cyclase 9 |
| chr11_-_88755360 | 1.79 |
ENSMUST00000018572.11
|
Akap1
|
A kinase (PRKA) anchor protein 1 |
| chr17_-_65920481 | 1.71 |
ENSMUST00000024897.10
|
Vapa
|
vesicle-associated membrane protein, associated protein A |
| chr18_-_62044871 | 1.71 |
ENSMUST00000166783.3
ENSMUST00000049378.15 |
Ablim3
|
actin binding LIM protein family, member 3 |
| chr16_-_44979013 | 1.68 |
ENSMUST00000023344.10
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr8_+_27532583 | 1.62 |
ENSMUST00000033875.10
ENSMUST00000209525.2 |
Plpbp
|
pyridoxal phosphate binding protein |
| chr9_+_77824646 | 1.61 |
ENSMUST00000034904.14
|
Elovl5
|
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
| chr17_-_27158514 | 1.60 |
ENSMUST00000114935.9
ENSMUST00000025027.10 |
Cuta
|
cutA divalent cation tolerance homolog |
| chr1_+_151220222 | 1.58 |
ENSMUST00000023918.13
ENSMUST00000111887.10 ENSMUST00000097543.8 |
Ivns1abp
|
influenza virus NS1A binding protein |
| chr2_+_25318642 | 1.58 |
ENSMUST00000102919.4
|
Abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr2_+_122065230 | 1.54 |
ENSMUST00000110551.4
|
Sord
|
sorbitol dehydrogenase |
| chr3_+_67490068 | 1.52 |
ENSMUST00000029344.10
|
Mfsd1
|
major facilitator superfamily domain containing 1 |
| chr17_+_74835417 | 1.51 |
ENSMUST00000182944.8
ENSMUST00000182597.8 ENSMUST00000182133.8 ENSMUST00000183224.8 |
Birc6
|
baculoviral IAP repeat-containing 6 |
| chr6_+_13069757 | 1.49 |
ENSMUST00000124234.8
ENSMUST00000142211.8 ENSMUST00000031556.14 |
Tmem106b
|
transmembrane protein 106B |
| chr8_+_56747613 | 1.48 |
ENSMUST00000034026.10
|
Hpgd
|
hydroxyprostaglandin dehydrogenase 15 (NAD) |
| chr18_+_10617858 | 1.45 |
ENSMUST00000235020.2
|
Snrpd1
|
small nuclear ribonucleoprotein D1 |
| chr11_-_96807233 | 1.44 |
ENSMUST00000130774.2
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr8_+_27532623 | 1.43 |
ENSMUST00000209856.2
ENSMUST00000098851.12 ENSMUST00000211393.2 ENSMUST00000211518.2 |
Plpbp
|
pyridoxal phosphate binding protein |
| chr15_-_96953823 | 1.36 |
ENSMUST00000023101.10
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr2_+_90677499 | 1.36 |
ENSMUST00000136872.8
ENSMUST00000150232.8 ENSMUST00000111467.4 |
Mtch2
|
mitochondrial carrier 2 |
| chr9_-_103243039 | 1.35 |
ENSMUST00000035484.11
|
Cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr17_+_74835290 | 1.32 |
ENSMUST00000180037.8
|
Birc6
|
baculoviral IAP repeat-containing 6 |
| chr2_+_31462659 | 1.32 |
ENSMUST00000113482.8
|
Fubp3
|
far upstream element (FUSE) binding protein 3 |
| chr11_-_50216426 | 1.31 |
ENSMUST00000179865.8
ENSMUST00000020637.9 |
Canx
|
calnexin |
| chr11_-_118460736 | 1.31 |
ENSMUST00000136551.3
|
Rbfox3
|
RNA binding protein, fox-1 homolog (C. elegans) 3 |
| chr1_+_5153300 | 1.30 |
ENSMUST00000044369.13
ENSMUST00000194676.6 ENSMUST00000192029.6 ENSMUST00000192698.3 |
Atp6v1h
|
ATPase, H+ transporting, lysosomal V1 subunit H |
| chr2_-_37593287 | 1.30 |
ENSMUST00000072186.12
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr11_+_70735751 | 1.29 |
ENSMUST00000177731.8
ENSMUST00000108533.10 ENSMUST00000081362.13 ENSMUST00000178245.2 |
Rabep1
|
rabaptin, RAB GTPase binding effector protein 1 |
| chr11_+_93776965 | 1.28 |
ENSMUST00000063718.11
ENSMUST00000107854.9 |
Mbtd1
|
mbt domain containing 1 |
| chr4_-_34882917 | 1.24 |
ENSMUST00000098163.9
ENSMUST00000047950.6 |
Zfp292
|
zinc finger protein 292 |
| chr16_-_44978929 | 1.24 |
ENSMUST00000181177.2
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr2_-_38816229 | 1.21 |
ENSMUST00000076275.11
ENSMUST00000142130.2 |
Nr6a1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr8_+_91635192 | 1.21 |
ENSMUST00000211403.2
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr4_+_97665992 | 1.21 |
ENSMUST00000107062.9
ENSMUST00000052018.12 ENSMUST00000107057.8 |
Nfia
|
nuclear factor I/A |
| chr5_-_120641658 | 1.19 |
ENSMUST00000031597.7
|
Plbd2
|
phospholipase B domain containing 2 |
| chr17_+_24971952 | 1.15 |
ENSMUST00000044922.8
ENSMUST00000234202.2 |
Hs3st6
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 6 |
| chr17_-_24428351 | 1.15 |
ENSMUST00000024931.6
|
Ntn3
|
netrin 3 |
| chr7_-_28441205 | 1.14 |
ENSMUST00000056078.9
|
Mrps12
|
mitochondrial ribosomal protein S12 |
| chr14_+_69409251 | 1.14 |
ENSMUST00000062437.10
|
Nkx2-6
|
NK2 homeobox 6 |
| chr12_+_95658987 | 1.14 |
ENSMUST00000057324.4
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr16_-_44978986 | 1.13 |
ENSMUST00000180636.8
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr15_-_79816785 | 1.13 |
ENSMUST00000089293.11
ENSMUST00000109616.9 |
Cbx7
|
chromobox 7 |
| chr7_+_15795735 | 1.12 |
ENSMUST00000209369.2
|
Zfp541
|
zinc finger protein 541 |
| chr1_+_75119419 | 1.12 |
ENSMUST00000097694.11
ENSMUST00000190240.7 |
Retreg2
|
reticulophagy regulator family member 2 |
| chr11_-_120508713 | 1.11 |
ENSMUST00000106188.4
ENSMUST00000026129.16 |
Pcyt2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr13_-_104246084 | 1.10 |
ENSMUST00000224945.2
ENSMUST00000109315.5 |
Nln
|
neurolysin (metallopeptidase M3 family) |
| chr11_+_60308077 | 1.10 |
ENSMUST00000070681.7
|
Gid4
|
GID complex subunit 4, VID24 homolog |
| chr7_-_28441173 | 1.09 |
ENSMUST00000171183.2
|
Mrps12
|
mitochondrial ribosomal protein S12 |
| chr1_-_175319842 | 1.09 |
ENSMUST00000195324.6
ENSMUST00000192227.6 ENSMUST00000194555.6 |
Rgs7
|
regulator of G protein signaling 7 |
| chr2_+_28531239 | 1.09 |
ENSMUST00000028155.12
ENSMUST00000113869.8 ENSMUST00000113867.9 |
Tsc1
|
TSC complex subunit 1 |
| chr10_+_127894816 | 1.08 |
ENSMUST00000052798.14
|
Ptges3
|
prostaglandin E synthase 3 |
| chr1_-_183078488 | 1.08 |
ENSMUST00000057062.12
|
Brox
|
BRO1 domain and CAAX motif containing |
| chr15_+_81695615 | 1.07 |
ENSMUST00000023024.8
|
Tef
|
thyrotroph embryonic factor |
| chr17_+_37253916 | 1.06 |
ENSMUST00000173072.2
|
Rnf39
|
ring finger protein 39 |
| chr10_+_127894843 | 1.06 |
ENSMUST00000084771.3
|
Ptges3
|
prostaglandin E synthase 3 |
| chr3_-_142587678 | 1.05 |
ENSMUST00000043812.15
|
Pkn2
|
protein kinase N2 |
| chr10_-_62322356 | 1.04 |
ENSMUST00000092473.5
|
Vps26a
|
VPS26 retromer complex component A |
| chr16_-_4867703 | 1.04 |
ENSMUST00000115844.3
ENSMUST00000023189.15 |
Glyr1
|
glyoxylate reductase 1 homolog (Arabidopsis) |
| chr13_-_112788890 | 1.03 |
ENSMUST00000099166.10
|
Ddx4
|
DEAD box helicase 4 |
| chr14_+_4230658 | 1.01 |
ENSMUST00000225491.2
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr19_-_60569323 | 1.00 |
ENSMUST00000111460.5
ENSMUST00000166712.9 ENSMUST00000081790.15 |
Cacul1
|
CDK2 associated, cullin domain 1 |
| chr1_+_84817547 | 0.99 |
ENSMUST00000097672.4
|
Fbxo36
|
F-box protein 36 |
| chr2_+_179666744 | 0.99 |
ENSMUST00000055485.12
|
Lsm14b
|
LSM family member 14B |
| chr13_-_112788829 | 0.99 |
ENSMUST00000075748.7
|
Ddx4
|
DEAD box helicase 4 |
| chr15_-_95426108 | 0.98 |
ENSMUST00000075275.3
|
Nell2
|
NEL-like 2 |
| chr1_+_75119472 | 0.98 |
ENSMUST00000189650.7
|
Retreg2
|
reticulophagy regulator family member 2 |
| chr12_-_59266511 | 0.98 |
ENSMUST00000043204.8
|
Fbxo33
|
F-box protein 33 |
| chr9_-_59657899 | 0.98 |
ENSMUST00000213257.2
ENSMUST00000216329.2 ENSMUST00000163586.9 ENSMUST00000217093.2 ENSMUST00000051039.5 ENSMUST00000177963.8 |
Senp8
|
SUMO/sentrin specific peptidase 8 |
| chr19_+_38825005 | 0.98 |
ENSMUST00000037302.6
|
Tbc1d12
|
TBC1D12: TBC1 domain family, member 12 |
| chr3_-_88997261 | 0.97 |
ENSMUST00000196223.5
ENSMUST00000196043.2 ENSMUST00000166687.6 |
Rusc1
|
RUN and SH3 domain containing 1 |
| chr11_-_17161504 | 0.96 |
ENSMUST00000020317.8
|
Pno1
|
partner of NOB1 homolog |
| chr13_+_9143995 | 0.96 |
ENSMUST00000091829.4
|
Larp4b
|
La ribonucleoprotein domain family, member 4B |
| chr5_+_77099154 | 0.95 |
ENSMUST00000031160.16
ENSMUST00000120912.8 ENSMUST00000117536.8 |
Paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoribosylaminoimidazole, succinocarboxamide synthetase |
| chr9_-_53617508 | 0.95 |
ENSMUST00000068449.4
|
Rab39
|
RAB39, member RAS oncogene family |
| chr9_-_103242737 | 0.94 |
ENSMUST00000072249.13
ENSMUST00000189896.2 |
Cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr1_-_75156993 | 0.93 |
ENSMUST00000027396.15
|
Abcb6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr17_-_36290571 | 0.92 |
ENSMUST00000173724.2
ENSMUST00000172900.8 ENSMUST00000174849.8 |
Prr3
|
proline-rich polypeptide 3 |
| chr7_+_44499818 | 0.92 |
ENSMUST00000136232.2
ENSMUST00000207223.2 |
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr17_-_80203457 | 0.92 |
ENSMUST00000068282.7
ENSMUST00000112437.8 |
Atl2
|
atlastin GTPase 2 |
| chr9_-_121686601 | 0.91 |
ENSMUST00000213124.2
ENSMUST00000215300.2 ENSMUST00000213147.2 |
Higd1a
|
HIG1 domain family, member 1A |
| chr13_+_38529062 | 0.90 |
ENSMUST00000171970.3
|
Bmp6
|
bone morphogenetic protein 6 |
| chr5_+_77099229 | 0.90 |
ENSMUST00000141687.2
|
Paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoribosylaminoimidazole, succinocarboxamide synthetase |
| chr11_+_70735572 | 0.90 |
ENSMUST00000076270.13
ENSMUST00000179114.8 ENSMUST00000100928.11 |
Rabep1
|
rabaptin, RAB GTPase binding effector protein 1 |
| chr10_-_115151451 | 0.89 |
ENSMUST00000020343.9
ENSMUST00000218831.2 |
Rab21
|
RAB21, member RAS oncogene family |
| chr4_-_148123223 | 0.89 |
ENSMUST00000030879.12
ENSMUST00000137724.8 |
Clcn6
|
chloride channel, voltage-sensitive 6 |
| chr9_-_91247831 | 0.88 |
ENSMUST00000065360.5
|
Zic1
|
zinc finger protein of the cerebellum 1 |
| chr17_+_46694646 | 0.88 |
ENSMUST00000113481.9
ENSMUST00000138127.8 |
Zfp318
|
zinc finger protein 318 |
| chr2_-_130266162 | 0.88 |
ENSMUST00000089581.11
|
Pced1a
|
PC-esterase domain containing 1A |
| chr1_+_75119809 | 0.88 |
ENSMUST00000186037.7
ENSMUST00000187901.2 |
Retreg2
|
reticulophagy regulator family member 2 |
| chr7_-_142522454 | 0.87 |
ENSMUST00000121862.3
|
Ascl2
|
achaete-scute family bHLH transcription factor 2 |
| chr15_-_79816717 | 0.87 |
ENSMUST00000177044.2
ENSMUST00000109615.8 |
Cbx7
|
chromobox 7 |
| chr2_-_6327884 | 0.85 |
ENSMUST00000238876.2
|
1700014B07Rik
|
RIKEN cDNA 1700014B07 gene |
| chr6_+_110622533 | 0.85 |
ENSMUST00000071076.13
ENSMUST00000172951.2 |
Grm7
|
glutamate receptor, metabotropic 7 |
| chr13_+_9143916 | 0.84 |
ENSMUST00000188211.8
ENSMUST00000188939.7 ENSMUST00000190041.7 |
Larp4b
|
La ribonucleoprotein domain family, member 4B |
| chr7_-_46445085 | 0.84 |
ENSMUST00000123725.2
|
Hps5
|
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr11_+_93776650 | 0.84 |
ENSMUST00000107853.8
ENSMUST00000107850.8 |
Mbtd1
|
mbt domain containing 1 |
| chr9_-_57513510 | 0.84 |
ENSMUST00000215487.2
ENSMUST00000045068.10 |
Cplx3
|
complexin 3 |
| chr9_-_91247809 | 0.83 |
ENSMUST00000034927.13
|
Zic1
|
zinc finger protein of the cerebellum 1 |
| chr19_+_38825120 | 0.83 |
ENSMUST00000235558.2
|
Tbc1d12
|
TBC1D12: TBC1 domain family, member 12 |
| chr1_+_182591771 | 0.83 |
ENSMUST00000193660.6
|
Susd4
|
sushi domain containing 4 |
| chr14_+_30741115 | 0.83 |
ENSMUST00000112094.8
ENSMUST00000144009.2 |
Pbrm1
|
polybromo 1 |
| chr19_-_42117420 | 0.83 |
ENSMUST00000161873.2
ENSMUST00000018965.4 |
Avpi1
|
arginine vasopressin-induced 1 |
| chrX_+_13147209 | 0.82 |
ENSMUST00000000804.7
|
Ddx3x
|
DEAD box helicase 3, X-linked |
| chr17_-_36290129 | 0.81 |
ENSMUST00000165613.9
ENSMUST00000173872.8 |
Prr3
|
proline-rich polypeptide 3 |
| chr14_-_31552335 | 0.81 |
ENSMUST00000228037.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr4_+_97660971 | 0.80 |
ENSMUST00000152023.8
|
Nfia
|
nuclear factor I/A |
| chr11_+_74540284 | 0.80 |
ENSMUST00000117818.2
ENSMUST00000092915.12 |
Cluh
|
clustered mitochondria (cluA/CLU1) homolog |
| chr2_-_48839218 | 0.80 |
ENSMUST00000090976.10
ENSMUST00000149679.8 |
Orc4
|
origin recognition complex, subunit 4 |
| chr8_+_26091607 | 0.79 |
ENSMUST00000155861.8
|
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr17_-_56933872 | 0.79 |
ENSMUST00000047226.10
|
Lonp1
|
lon peptidase 1, mitochondrial |
| chr5_-_149559792 | 0.78 |
ENSMUST00000202361.4
ENSMUST00000202089.4 ENSMUST00000200825.2 ENSMUST00000201559.4 |
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr12_-_3359924 | 0.78 |
ENSMUST00000021001.10
|
Rab10
|
RAB10, member RAS oncogene family |
| chrX_+_138511360 | 0.77 |
ENSMUST00000113026.2
|
Rnf128
|
ring finger protein 128 |
| chr2_+_37342248 | 0.77 |
ENSMUST00000148470.8
ENSMUST00000066055.10 ENSMUST00000112920.2 |
Rabgap1
|
RAB GTPase activating protein 1 |
| chr2_-_48839276 | 0.76 |
ENSMUST00000028098.11
|
Orc4
|
origin recognition complex, subunit 4 |
| chr16_-_44978546 | 0.76 |
ENSMUST00000114600.2
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr10_-_62322451 | 0.75 |
ENSMUST00000217868.2
|
Vps26a
|
VPS26 retromer complex component A |
| chr19_+_41900360 | 0.75 |
ENSMUST00000011896.8
|
Pgam1
|
phosphoglycerate mutase 1 |
| chr9_-_20887967 | 0.75 |
ENSMUST00000214218.2
|
S1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr2_+_19450443 | 0.75 |
ENSMUST00000028068.3
|
Ptf1a
|
pancreas specific transcription factor, 1a |
| chr3_+_94861386 | 0.74 |
ENSMUST00000107260.9
ENSMUST00000142311.8 ENSMUST00000137088.8 ENSMUST00000152869.8 ENSMUST00000107254.8 ENSMUST00000107253.8 |
Rfx5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr11_+_117545037 | 0.74 |
ENSMUST00000026658.13
|
Tnrc6c
|
trinucleotide repeat containing 6C |
| chr6_-_83433357 | 0.74 |
ENSMUST00000186548.7
|
Tet3
|
tet methylcytosine dioxygenase 3 |
| chr3_-_88204145 | 0.73 |
ENSMUST00000010682.4
|
Tsacc
|
TSSK6 activating co-chaperone |
| chr5_-_45796857 | 0.73 |
ENSMUST00000016023.9
|
Fam184b
|
family with sequence similarity 184, member B |
| chr16_-_44978816 | 0.73 |
ENSMUST00000181750.2
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr5_-_52827015 | 0.72 |
ENSMUST00000031069.13
|
Sepsecs
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr11_-_51891259 | 0.71 |
ENSMUST00000020657.13
|
Ube2b
|
ubiquitin-conjugating enzyme E2B |
| chr3_-_88204286 | 0.70 |
ENSMUST00000107556.10
|
Tsacc
|
TSSK6 activating co-chaperone |
| chr11_-_102588536 | 0.70 |
ENSMUST00000164506.3
ENSMUST00000092569.13 |
Ccdc43
|
coiled-coil domain containing 43 |
| chr14_+_26300693 | 0.69 |
ENSMUST00000203874.3
ENSMUST00000037585.9 |
Dennd6a
|
DENN/MADD domain containing 6A |
| chr9_-_123546605 | 0.69 |
ENSMUST00000163207.2
|
Lztfl1
|
leucine zipper transcription factor-like 1 |
| chr17_+_25352353 | 0.68 |
ENSMUST00000162862.3
ENSMUST00000040729.9 |
Clcn7
|
chloride channel, voltage-sensitive 7 |
| chr2_+_31462780 | 0.68 |
ENSMUST00000137889.7
ENSMUST00000194386.6 ENSMUST00000055244.13 |
Fubp3
|
far upstream element (FUSE) binding protein 3 |
| chr13_-_48779047 | 0.68 |
ENSMUST00000222028.2
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chr9_-_123546642 | 0.68 |
ENSMUST00000165754.8
ENSMUST00000026274.14 |
Lztfl1
|
leucine zipper transcription factor-like 1 |
| chr11_+_57899890 | 0.68 |
ENSMUST00000071487.13
ENSMUST00000178636.2 |
Larp1
|
La ribonucleoprotein domain family, member 1 |
| chr6_+_94477294 | 0.67 |
ENSMUST00000061118.11
|
Slc25a26
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 26 |
| chr1_+_87254729 | 0.67 |
ENSMUST00000172794.8
ENSMUST00000164992.9 ENSMUST00000173173.8 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr16_+_93885775 | 0.66 |
ENSMUST00000072182.9
|
Sim2
|
single-minded family bHLH transcription factor 2 |
| chr2_+_83474779 | 0.66 |
ENSMUST00000081591.7
|
Zc3h15
|
zinc finger CCCH-type containing 15 |
| chr10_+_121200984 | 0.65 |
ENSMUST00000040344.7
|
Gns
|
glucosamine (N-acetyl)-6-sulfatase |
| chr13_-_54835460 | 0.65 |
ENSMUST00000129881.8
|
Rnf44
|
ring finger protein 44 |
| chr3_+_108093645 | 0.65 |
ENSMUST00000050909.7
ENSMUST00000106659.3 ENSMUST00000106656.2 |
Amigo1
|
adhesion molecule with Ig like domain 1 |
| chr11_+_117545618 | 0.65 |
ENSMUST00000106344.8
|
Tnrc6c
|
trinucleotide repeat containing 6C |
| chr13_+_104246245 | 0.64 |
ENSMUST00000044385.14
|
Sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr14_+_30741082 | 0.64 |
ENSMUST00000112098.11
ENSMUST00000112095.8 ENSMUST00000112106.8 ENSMUST00000146325.8 |
Pbrm1
|
polybromo 1 |
| chr18_+_14916295 | 0.63 |
ENSMUST00000234789.2
ENSMUST00000169862.2 |
Taf4b
|
TATA-box binding protein associated factor 4b |
| chr6_+_29768470 | 0.63 |
ENSMUST00000102995.9
ENSMUST00000115242.9 |
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr7_+_141035111 | 0.63 |
ENSMUST00000164016.8
ENSMUST00000064151.13 ENSMUST00000169665.8 |
Pnpla2
|
patatin-like phospholipase domain containing 2 |
| chr17_-_56916771 | 0.63 |
ENSMUST00000052832.6
|
Micos13
|
mitochondrial contact site and cristae organizing system subunit 13 |
| chr10_+_95776543 | 0.62 |
ENSMUST00000053484.8
|
Eea1
|
early endosome antigen 1 |
| chr2_-_126718037 | 0.62 |
ENSMUST00000028843.12
|
Trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr8_+_80366247 | 0.61 |
ENSMUST00000173078.8
ENSMUST00000173286.8 |
Otud4
|
OTU domain containing 4 |
| chr2_+_48839505 | 0.61 |
ENSMUST00000112745.8
ENSMUST00000112754.8 |
Mbd5
|
methyl-CpG binding domain protein 5 |
| chr16_-_10884005 | 0.61 |
ENSMUST00000162323.2
|
Litaf
|
LPS-induced TN factor |
| chr17_-_5468938 | 0.61 |
ENSMUST00000189788.2
|
Ldhal6b
|
lactate dehydrogenase A-like 6B |
| chr16_+_8647959 | 0.60 |
ENSMUST00000023150.7
|
1810013L24Rik
|
RIKEN cDNA 1810013L24 gene |
| chr11_+_100978103 | 0.58 |
ENSMUST00000107302.8
ENSMUST00000107303.10 ENSMUST00000017945.15 ENSMUST00000149597.2 |
Mlx
|
MAX-like protein X |
| chr11_-_104333059 | 0.58 |
ENSMUST00000106977.8
ENSMUST00000106972.8 |
Kansl1
|
KAT8 regulatory NSL complex subunit 1 |
| chr8_-_70426910 | 0.58 |
ENSMUST00000116463.4
|
Gatad2a
|
GATA zinc finger domain containing 2A |
| chr6_+_64706101 | 0.58 |
ENSMUST00000101351.6
|
Atoh1
|
atonal bHLH transcription factor 1 |
| chr3_+_96634974 | 0.58 |
ENSMUST00000029740.14
|
Rnf115
|
ring finger protein 115 |
| chr13_-_48779072 | 0.58 |
ENSMUST00000035824.11
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chr17_+_34866160 | 0.58 |
ENSMUST00000173984.2
|
Atf6b
|
activating transcription factor 6 beta |
| chr6_-_99703344 | 0.57 |
ENSMUST00000008273.8
ENSMUST00000101120.11 ENSMUST00000203738.2 |
Prok2
|
prokineticin 2 |
| chr10_+_97528915 | 0.57 |
ENSMUST00000060703.6
|
Ccer1
|
coiled-coil glutamate-rich protein 1 |
| chr3_+_89680867 | 0.57 |
ENSMUST00000038356.13
|
Ube2q1
|
ubiquitin-conjugating enzyme E2Q family member 1 |
| chr2_+_146063841 | 0.57 |
ENSMUST00000089257.6
|
Insm1
|
insulinoma-associated 1 |
| chr3_+_107502347 | 0.56 |
ENSMUST00000014747.3
|
Alx3
|
aristaless-like homeobox 3 |
| chr7_+_127111576 | 0.56 |
ENSMUST00000186672.7
|
Srcap
|
Snf2-related CREBBP activator protein |
| chr11_+_118913788 | 0.56 |
ENSMUST00000026662.8
|
Cbx2
|
chromobox 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Arnt
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 1.0 | 17.4 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.8 | 2.4 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.6 | 3.2 | GO:1904253 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.6 | 3.9 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.5 | 1.5 | GO:0006059 | hexitol metabolic process(GO:0006059) alditol catabolic process(GO:0019405) |
| 0.5 | 2.5 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.4 | 2.6 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.4 | 1.1 | GO:0090649 | rRNA export from nucleus(GO:0006407) response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.3 | 2.4 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.3 | 1.1 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.3 | 6.8 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.3 | 0.8 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.3 | 0.8 | GO:0060466 | activation of meiosis involved in egg activation(GO:0060466) |
| 0.3 | 0.8 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.3 | 2.0 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.3 | 0.5 | GO:0033128 | negative regulation of histone phosphorylation(GO:0033128) |
| 0.2 | 1.6 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.2 | 0.7 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.2 | 1.7 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.2 | 0.8 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 0.2 | 1.8 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.2 | 1.2 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.2 | 1.4 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.2 | 0.9 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.2 | 1.4 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.2 | 0.7 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.2 | 5.8 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.2 | 2.2 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.2 | 0.5 | GO:1902868 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.2 | 1.1 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.2 | 2.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.2 | 0.6 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 0.4 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.1 | 0.7 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.1 | 0.4 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.9 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.6 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.1 | 0.8 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 3.9 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 1.4 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.1 | 0.9 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.1 | 1.6 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.9 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 2.8 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.1 | 0.9 | GO:0032349 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.1 | 0.4 | GO:0070429 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.1 | 1.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 2.1 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 0.8 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 0.5 | GO:0071895 | negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) |
| 0.1 | 0.1 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 1.2 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.1 | 0.5 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.8 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 1.5 | GO:0007567 | parturition(GO:0007567) |
| 0.1 | 0.3 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.1 | 0.7 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.1 | 0.9 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.6 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 0.3 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 2.0 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 0.7 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 | 0.6 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 2.1 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 1.3 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 1.4 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 0.5 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.1 | 2.0 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 0.9 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.1 | 2.0 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.4 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 0.2 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 | 1.1 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 4.5 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 | 2.9 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.1 | 1.8 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.1 | 0.1 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.1 | 0.7 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.2 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.1 | 1.0 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.6 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.2 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.1 | 1.0 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 0.6 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.1 | 0.2 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 2.3 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 0.4 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 0.6 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.1 | 0.2 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.1 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.2 | GO:0002660 | CD4-positive, CD25-positive, alpha-beta regulatory T cell lineage commitment(GO:0002362) positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.1 | 0.4 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 2.7 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.1 | 0.4 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 0.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.4 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.6 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.4 | GO:0051126 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) negative regulation of actin nucleation(GO:0051126) |
| 0.0 | 0.8 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.6 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.2 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 0.5 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.5 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 1.6 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 1.6 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.2 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.9 | GO:0090201 | negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.0 | 2.2 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 1.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.0 | 0.6 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.6 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 1.7 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.0 | 0.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 1.6 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.1 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.4 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.0 | 1.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 5.4 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.6 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.6 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 1.6 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 1.6 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 1.2 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.1 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) response to methylglyoxal(GO:0051595) regulation of histone H3-K27 acetylation(GO:1901674) negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.8 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.8 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 1.2 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.5 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 1.2 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 2.1 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.3 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.9 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.1 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.0 | 1.6 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 | 0.3 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 0.5 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.0 | 0.7 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.3 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.0 | 1.3 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.1 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 1.6 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 18.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 2.0 | 5.9 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.8 | 3.9 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.4 | 1.6 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.4 | 1.5 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.3 | 2.0 | GO:0071547 | piP-body(GO:0071547) |
| 0.3 | 1.3 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.3 | 1.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.3 | 1.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.3 | 2.4 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.2 | 2.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.2 | 2.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 2.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.4 | GO:0005712 | chiasma(GO:0005712) |
| 0.1 | 1.6 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.0 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 0.6 | GO:0044308 | axonal spine(GO:0044308) |
| 0.1 | 0.5 | GO:1990590 | Lewy body core(GO:1990037) ATF4-CREB1 transcription factor complex(GO:1990589) ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.1 | 2.4 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.1 | 2.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 1.3 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.5 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 1.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.3 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 0.7 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 1.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 2.2 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 0.5 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.8 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 1.9 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 1.1 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 1.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.3 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 1.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.5 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.8 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.8 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 1.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 5.7 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 2.6 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 2.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.8 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.4 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.6 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.6 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.5 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 2.5 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.9 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 3.3 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 4.7 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.9 | GO:0070469 | respiratory chain(GO:0070469) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.6 | 2.4 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.6 | 1.7 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.5 | 2.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.4 | 2.6 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.3 | 1.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.3 | 0.8 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.2 | 2.9 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.2 | 1.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.2 | 5.5 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.2 | 0.8 | GO:0070905 | serine binding(GO:0070905) |
| 0.2 | 1.5 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.2 | 0.8 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.2 | 1.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 0.9 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.4 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 1.0 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.7 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 1.6 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.8 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 1.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 2.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 3.5 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 0.4 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 1.8 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 0.3 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.1 | 0.4 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 0.7 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 2.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.8 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.1 | 2.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.7 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.6 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 1.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 2.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.6 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 1.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 3.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.4 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 1.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 5.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.5 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.1 | 2.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 4.1 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 1.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 0.7 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.6 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 4.5 | GO:0051287 | NAD binding(GO:0051287) |
| 0.1 | 0.6 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 4.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.3 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 1.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 1.9 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 1.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.5 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.2 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 1.7 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 1.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 2.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.6 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.8 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 3.1 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 1.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.8 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0052692 | raffinose alpha-galactosidase activity(GO:0052692) |
| 0.0 | 0.8 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 1.9 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 6.6 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.3 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 1.9 | GO:0008186 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 1.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.5 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.6 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 1.8 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.8 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 2.3 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.2 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 1.4 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.0 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 3.5 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 2.8 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 2.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 2.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 2.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 3.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.6 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.3 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 4.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.6 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 1.8 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 2.1 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 3.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 2.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.4 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 1.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 0.7 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 2.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 0.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.1 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 4.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.4 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.9 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.5 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.8 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 1.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 2.1 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 1.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.1 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |