Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
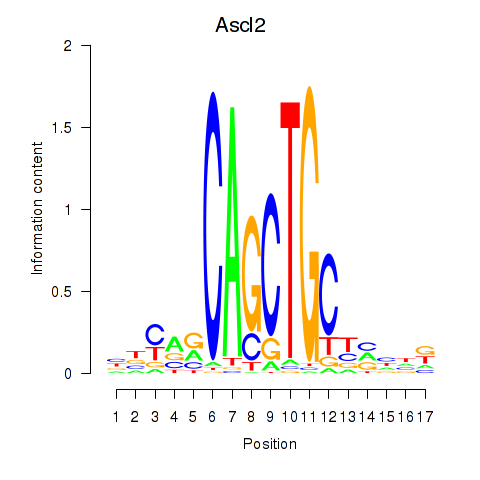
Results for Ascl2
Z-value: 0.90
Transcription factors associated with Ascl2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ascl2
|
ENSMUSG00000009248.7 | achaete-scute family bHLH transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ascl2 | mm39_v1_chr7_-_142522995_142523006 | 0.55 | 4.8e-04 | Click! |
Activity profile of Ascl2 motif
Sorted Z-values of Ascl2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_112417633 | 8.57 |
ENSMUST00000034435.7
|
Ctrb1
|
chymotrypsinogen B1 |
| chr7_+_44866635 | 8.22 |
ENSMUST00000097216.5
ENSMUST00000209343.2 ENSMUST00000209678.2 |
Tead2
|
TEA domain family member 2 |
| chr7_-_142223662 | 7.95 |
ENSMUST00000228850.2
|
Gm49394
|
predicted gene, 49394 |
| chr10_-_128237087 | 6.95 |
ENSMUST00000042666.13
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr9_+_110867807 | 5.84 |
ENSMUST00000197575.2
|
Ltf
|
lactotransferrin |
| chr19_-_15901919 | 5.61 |
ENSMUST00000162053.8
|
Psat1
|
phosphoserine aminotransferase 1 |
| chr15_-_74983786 | 5.32 |
ENSMUST00000191451.2
ENSMUST00000100542.10 |
Ly6c2
|
lymphocyte antigen 6 complex, locus C2 |
| chr7_-_45173193 | 5.05 |
ENSMUST00000211212.2
|
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr8_+_95703728 | 4.88 |
ENSMUST00000179619.9
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr4_-_137157824 | 4.73 |
ENSMUST00000102522.5
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr4_-_133600308 | 4.38 |
ENSMUST00000137486.3
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr11_+_53410697 | 4.24 |
ENSMUST00000120878.9
ENSMUST00000147912.2 |
Septin8
|
septin 8 |
| chr3_+_107803225 | 4.03 |
ENSMUST00000172247.8
ENSMUST00000167387.8 |
Gstm5
|
glutathione S-transferase, mu 5 |
| chr3_-_100396635 | 3.84 |
ENSMUST00000061455.9
|
Tent5c
|
terminal nucleotidyltransferase 5C |
| chr17_-_35285146 | 3.83 |
ENSMUST00000174190.2
ENSMUST00000097337.8 |
Mpig6b
|
megakaryocyte and platelet inhibitory receptor G6b |
| chr13_+_54849268 | 3.69 |
ENSMUST00000037145.8
|
Cdhr2
|
cadherin-related family member 2 |
| chr9_-_21874802 | 3.57 |
ENSMUST00000006397.7
|
Epor
|
erythropoietin receptor |
| chr7_+_142025575 | 3.44 |
ENSMUST00000038946.9
|
Lsp1
|
lymphocyte specific 1 |
| chr3_+_107803137 | 3.33 |
ENSMUST00000004134.11
|
Gstm5
|
glutathione S-transferase, mu 5 |
| chr1_-_167221344 | 3.31 |
ENSMUST00000028005.3
|
Mgst3
|
microsomal glutathione S-transferase 3 |
| chr12_-_113386312 | 3.25 |
ENSMUST00000177715.8
ENSMUST00000103426.3 |
Ighm
|
immunoglobulin heavy constant mu |
| chr7_+_44866095 | 3.19 |
ENSMUST00000209437.2
|
Tead2
|
TEA domain family member 2 |
| chr11_-_46203047 | 3.14 |
ENSMUST00000129474.2
ENSMUST00000093166.11 ENSMUST00000165599.9 |
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr7_+_142025817 | 3.06 |
ENSMUST00000105966.2
|
Lsp1
|
lymphocyte specific 1 |
| chr17_+_56259617 | 3.06 |
ENSMUST00000003274.8
|
Ebi3
|
Epstein-Barr virus induced gene 3 |
| chr7_+_78563964 | 3.04 |
ENSMUST00000120331.4
|
Isg20
|
interferon-stimulated protein |
| chr2_+_84564394 | 2.94 |
ENSMUST00000238573.2
ENSMUST00000090729.9 |
Ypel4
|
yippee like 4 |
| chr7_+_78564062 | 2.85 |
ENSMUST00000205981.2
|
Isg20
|
interferon-stimulated protein |
| chr4_-_63965161 | 2.80 |
ENSMUST00000107377.10
|
Tnc
|
tenascin C |
| chr11_+_104468107 | 2.79 |
ENSMUST00000106956.10
|
Myl4
|
myosin, light polypeptide 4 |
| chr6_+_29694181 | 2.79 |
ENSMUST00000046750.14
ENSMUST00000115250.4 |
Tspan33
|
tetraspanin 33 |
| chr1_+_87603952 | 2.74 |
ENSMUST00000170300.8
ENSMUST00000167032.2 |
Inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr11_+_98798627 | 2.74 |
ENSMUST00000092706.13
|
Cdc6
|
cell division cycle 6 |
| chr1_-_88133472 | 2.69 |
ENSMUST00000119972.4
|
Dnajb3
|
DnaJ heat shock protein family (Hsp40) member B3 |
| chr11_+_104467791 | 2.67 |
ENSMUST00000106957.8
|
Myl4
|
myosin, light polypeptide 4 |
| chrX_-_36253309 | 2.64 |
ENSMUST00000060474.14
ENSMUST00000053456.11 ENSMUST00000115239.10 |
Septin6
|
septin 6 |
| chr11_+_117740077 | 2.62 |
ENSMUST00000081387.11
|
Birc5
|
baculoviral IAP repeat-containing 5 |
| chr7_-_141023199 | 2.59 |
ENSMUST00000106005.9
|
Pidd1
|
p53 induced death domain protein 1 |
| chr16_-_17394495 | 2.54 |
ENSMUST00000231645.2
ENSMUST00000232226.2 ENSMUST00000232336.2 ENSMUST00000232385.2 ENSMUST00000231615.2 ENSMUST00000231283.2 ENSMUST00000172164.10 |
Slc7a4
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 4 |
| chr18_-_32692967 | 2.50 |
ENSMUST00000174000.2
ENSMUST00000174459.2 |
Gypc
|
glycophorin C |
| chr9_+_107852733 | 2.47 |
ENSMUST00000035216.11
|
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr1_+_174000304 | 2.46 |
ENSMUST00000027817.8
|
Spta1
|
spectrin alpha, erythrocytic 1 |
| chr2_-_150510116 | 2.33 |
ENSMUST00000028944.4
|
Acss1
|
acyl-CoA synthetase short-chain family member 1 |
| chr8_+_95703506 | 2.30 |
ENSMUST00000212581.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr3_+_107803563 | 2.25 |
ENSMUST00000169365.2
|
Gstm5
|
glutathione S-transferase, mu 5 |
| chr7_-_127593003 | 2.22 |
ENSMUST00000033056.5
|
Pycard
|
PYD and CARD domain containing |
| chr13_+_95833359 | 2.22 |
ENSMUST00000022182.5
|
F2rl2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr4_-_133599616 | 2.21 |
ENSMUST00000157067.9
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr7_-_80037153 | 2.19 |
ENSMUST00000206728.2
|
Fes
|
feline sarcoma oncogene |
| chr17_+_57586094 | 2.17 |
ENSMUST00000169220.9
ENSMUST00000005889.16 ENSMUST00000112870.5 |
Vav1
|
vav 1 oncogene |
| chr3_+_146110387 | 2.06 |
ENSMUST00000106151.8
ENSMUST00000106153.9 ENSMUST00000039021.11 ENSMUST00000106149.8 ENSMUST00000149262.8 |
Ssx2ip
|
synovial sarcoma, X 2 interacting protein |
| chr14_+_33645539 | 2.03 |
ENSMUST00000168727.3
|
Gdf10
|
growth differentiation factor 10 |
| chr15_+_75027089 | 2.01 |
ENSMUST00000190262.2
|
Ly6g
|
lymphocyte antigen 6 complex, locus G |
| chr7_+_142014546 | 2.01 |
ENSMUST00000105968.8
ENSMUST00000018963.11 ENSMUST00000105967.8 |
Lsp1
|
lymphocyte specific 1 |
| chr8_+_23629080 | 1.99 |
ENSMUST00000033947.15
|
Ank1
|
ankyrin 1, erythroid |
| chr11_+_117740111 | 1.97 |
ENSMUST00000093906.5
|
Birc5
|
baculoviral IAP repeat-containing 5 |
| chr14_-_56181993 | 1.95 |
ENSMUST00000022834.7
ENSMUST00000226280.2 |
Cma1
|
chymase 1, mast cell |
| chr13_+_97377604 | 1.94 |
ENSMUST00000041623.9
|
Enc1
|
ectodermal-neural cortex 1 |
| chr5_+_136996713 | 1.90 |
ENSMUST00000001790.6
|
Cldn15
|
claudin 15 |
| chr4_-_131695135 | 1.87 |
ENSMUST00000146443.8
ENSMUST00000135579.8 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr16_+_57173456 | 1.84 |
ENSMUST00000159816.8
|
Filip1l
|
filamin A interacting protein 1-like |
| chr8_-_41827639 | 1.82 |
ENSMUST00000034000.15
ENSMUST00000143057.2 |
Asah1
|
N-acylsphingosine amidohydrolase 1 |
| chr11_-_107607343 | 1.80 |
ENSMUST00000021065.6
|
Cacng1
|
calcium channel, voltage-dependent, gamma subunit 1 |
| chr11_+_83007635 | 1.79 |
ENSMUST00000037994.8
|
Slfn1
|
schlafen 1 |
| chr7_-_127805518 | 1.76 |
ENSMUST00000033049.9
|
Cox6a2
|
cytochrome c oxidase subunit 6A2 |
| chr15_-_78657640 | 1.75 |
ENSMUST00000018313.6
|
Mfng
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr17_-_26420300 | 1.75 |
ENSMUST00000025019.9
|
Arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr2_+_35172392 | 1.73 |
ENSMUST00000028239.8
|
Gsn
|
gelsolin |
| chr15_-_66703471 | 1.72 |
ENSMUST00000164163.8
|
Sla
|
src-like adaptor |
| chr5_+_123214332 | 1.72 |
ENSMUST00000067505.15
ENSMUST00000111619.10 ENSMUST00000160344.2 |
Tmem120b
|
transmembrane protein 120B |
| chrX_-_72954835 | 1.69 |
ENSMUST00000114404.8
ENSMUST00000114407.9 ENSMUST00000114406.9 ENSMUST00000064376.13 ENSMUST00000114405.8 |
Arhgap4
|
Rho GTPase activating protein 4 |
| chr19_-_6014210 | 1.68 |
ENSMUST00000025752.15
ENSMUST00000165143.3 |
Pola2
|
polymerase (DNA directed), alpha 2 |
| chr2_+_69652714 | 1.67 |
ENSMUST00000053087.4
|
Klhl23
|
kelch-like 23 |
| chr1_-_156501860 | 1.62 |
ENSMUST00000188964.7
ENSMUST00000190607.2 ENSMUST00000079625.11 |
Tor3a
|
torsin family 3, member A |
| chr1_+_172327812 | 1.62 |
ENSMUST00000192460.2
|
Tagln2
|
transgelin 2 |
| chrX_+_74425990 | 1.61 |
ENSMUST00000033541.5
|
Fundc2
|
FUN14 domain containing 2 |
| chr7_-_44180700 | 1.60 |
ENSMUST00000205506.2
|
Spib
|
Spi-B transcription factor (Spi-1/PU.1 related) |
| chr2_-_122441719 | 1.58 |
ENSMUST00000028624.9
|
Gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr3_-_129763638 | 1.56 |
ENSMUST00000146340.2
ENSMUST00000153506.8 |
Mcub
|
mitochondrial calcium uniporter dominant negative beta subunit |
| chr4_-_43523388 | 1.53 |
ENSMUST00000107913.10
ENSMUST00000030184.12 |
Tpm2
|
tropomyosin 2, beta |
| chr3_-_129763801 | 1.53 |
ENSMUST00000029624.15
|
Mcub
|
mitochondrial calcium uniporter dominant negative beta subunit |
| chr8_+_23629046 | 1.52 |
ENSMUST00000121075.8
|
Ank1
|
ankyrin 1, erythroid |
| chrX_-_105884178 | 1.52 |
ENSMUST00000062010.10
|
Rtl3
|
retrotransposon Gag like 3 |
| chr13_-_56326695 | 1.51 |
ENSMUST00000225063.2
|
Tifab
|
TRAF-interacting protein with forkhead-associated domain, family member B |
| chr11_+_69806866 | 1.51 |
ENSMUST00000134581.2
|
Gps2
|
G protein pathway suppressor 2 |
| chr3_+_108272205 | 1.49 |
ENSMUST00000090563.7
|
Mybphl
|
myosin binding protein H-like |
| chr12_+_26519203 | 1.48 |
ENSMUST00000020969.5
|
Cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr17_+_47983587 | 1.45 |
ENSMUST00000152724.2
|
Usp49
|
ubiquitin specific peptidase 49 |
| chr5_+_137639538 | 1.45 |
ENSMUST00000177466.8
ENSMUST00000166099.3 |
Sap25
|
sin3 associated polypeptide |
| chr19_-_6014159 | 1.44 |
ENSMUST00000235224.2
|
Pola2
|
polymerase (DNA directed), alpha 2 |
| chr15_+_78481247 | 1.44 |
ENSMUST00000043069.6
ENSMUST00000231180.2 ENSMUST00000229796.2 ENSMUST00000229295.2 |
Cyth4
|
cytohesin 4 |
| chr2_+_163916042 | 1.41 |
ENSMUST00000018353.14
|
Stk4
|
serine/threonine kinase 4 |
| chr4_-_43523595 | 1.40 |
ENSMUST00000107914.10
|
Tpm2
|
tropomyosin 2, beta |
| chr11_-_76289888 | 1.40 |
ENSMUST00000021204.4
|
Nxn
|
nucleoredoxin |
| chr2_+_32253692 | 1.38 |
ENSMUST00000113331.8
ENSMUST00000113338.9 |
Ciz1
|
CDKN1A interacting zinc finger protein 1 |
| chr13_-_56326511 | 1.37 |
ENSMUST00000169652.3
|
Tifab
|
TRAF-interacting protein with forkhead-associated domain, family member B |
| chr7_+_3339077 | 1.35 |
ENSMUST00000203566.3
|
Myadm
|
myeloid-associated differentiation marker |
| chr12_-_114752425 | 1.34 |
ENSMUST00000103510.2
|
Ighv1-26
|
immunoglobulin heavy variable 1-26 |
| chr9_-_57743989 | 1.33 |
ENSMUST00000164010.8
ENSMUST00000171444.8 ENSMUST00000098686.4 |
Arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr7_+_3339059 | 1.32 |
ENSMUST00000096744.8
|
Myadm
|
myeloid-associated differentiation marker |
| chr6_+_135339543 | 1.32 |
ENSMUST00000205156.3
|
Emp1
|
epithelial membrane protein 1 |
| chr17_-_26420332 | 1.32 |
ENSMUST00000121959.3
|
Arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr7_-_80037622 | 1.30 |
ENSMUST00000206698.2
|
Fes
|
feline sarcoma oncogene |
| chrX_+_20483742 | 1.30 |
ENSMUST00000115375.8
ENSMUST00000115374.8 ENSMUST00000084383.10 |
Rbm10
|
RNA binding motif protein 10 |
| chr8_-_106198112 | 1.25 |
ENSMUST00000014990.13
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr12_+_73333641 | 1.25 |
ENSMUST00000153941.8
ENSMUST00000122920.8 ENSMUST00000101313.4 |
Slc38a6
|
solute carrier family 38, member 6 |
| chr7_+_120442048 | 1.20 |
ENSMUST00000047875.16
|
Eef2k
|
eukaryotic elongation factor-2 kinase |
| chr2_-_25086810 | 1.19 |
ENSMUST00000081869.7
|
Tor4a
|
torsin family 4, member A |
| chr11_+_117877118 | 1.18 |
ENSMUST00000132676.8
|
Pgs1
|
phosphatidylglycerophosphate synthase 1 |
| chr1_-_133352115 | 1.17 |
ENSMUST00000153799.8
|
Sox13
|
SRY (sex determining region Y)-box 13 |
| chr2_+_25152600 | 1.17 |
ENSMUST00000114336.4
|
Tprn
|
taperin |
| chr19_+_5118103 | 1.13 |
ENSMUST00000070630.8
|
Cd248
|
CD248 antigen, endosialin |
| chr13_+_55593116 | 1.11 |
ENSMUST00000001115.16
ENSMUST00000224995.2 ENSMUST00000225925.2 ENSMUST00000099482.5 ENSMUST00000224118.2 |
Grk6
|
G protein-coupled receptor kinase 6 |
| chr17_+_23945310 | 1.10 |
ENSMUST00000024701.9
|
Pkmyt1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr7_+_27259895 | 1.09 |
ENSMUST00000187032.2
|
2310022A10Rik
|
RIKEN cDNA 2310022A10 gene |
| chr17_+_48571298 | 1.09 |
ENSMUST00000059873.14
ENSMUST00000154335.8 ENSMUST00000136272.8 ENSMUST00000125426.8 ENSMUST00000153420.2 |
Treml4
|
triggering receptor expressed on myeloid cells-like 4 |
| chr2_-_93164812 | 1.08 |
ENSMUST00000111265.9
|
Tspan18
|
tetraspanin 18 |
| chr4_-_43523745 | 1.08 |
ENSMUST00000150592.2
|
Tpm2
|
tropomyosin 2, beta |
| chr8_+_70826191 | 1.08 |
ENSMUST00000003659.9
|
Comp
|
cartilage oligomeric matrix protein |
| chr1_-_182929025 | 1.06 |
ENSMUST00000171366.7
|
Disp1
|
dispatched RND transporter family member 1 |
| chr11_+_69856222 | 1.06 |
ENSMUST00000018713.13
|
Cldn7
|
claudin 7 |
| chr1_+_134383247 | 1.04 |
ENSMUST00000112232.8
ENSMUST00000027725.11 ENSMUST00000116528.2 |
Klhl12
|
kelch-like 12 |
| chr9_-_62444318 | 1.00 |
ENSMUST00000048043.12
|
Coro2b
|
coronin, actin binding protein, 2B |
| chr15_+_101310283 | 1.00 |
ENSMUST00000068904.9
|
Krt7
|
keratin 7 |
| chr17_-_34170401 | 1.00 |
ENSMUST00000087543.5
|
B3galt4
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 |
| chr2_-_25086770 | 0.96 |
ENSMUST00000142857.2
ENSMUST00000137920.2 |
Tor4a
|
torsin family 4, member A |
| chr7_+_29683373 | 0.95 |
ENSMUST00000148442.8
|
Zfp568
|
zinc finger protein 568 |
| chr15_-_96540760 | 0.95 |
ENSMUST00000088452.11
|
Slc38a1
|
solute carrier family 38, member 1 |
| chr11_+_121593219 | 0.95 |
ENSMUST00000036742.14
|
Metrnl
|
meteorin, glial cell differentiation regulator-like |
| chr6_+_135339929 | 0.94 |
ENSMUST00000032330.16
|
Emp1
|
epithelial membrane protein 1 |
| chr11_-_115426618 | 0.94 |
ENSMUST00000121185.8
ENSMUST00000117589.8 |
Sumo2
|
small ubiquitin-like modifier 2 |
| chr6_-_85351524 | 0.94 |
ENSMUST00000060837.10
|
Rab11fip5
|
RAB11 family interacting protein 5 (class I) |
| chr7_+_28488380 | 0.92 |
ENSMUST00000209035.2
ENSMUST00000059857.8 |
Rinl
|
Ras and Rab interactor-like |
| chr4_+_140633646 | 0.92 |
ENSMUST00000030765.7
|
Padi2
|
peptidyl arginine deiminase, type II |
| chr9_+_107784065 | 0.90 |
ENSMUST00000035203.9
|
Mst1r
|
macrophage stimulating 1 receptor (c-met-related tyrosine kinase) |
| chr2_+_164782642 | 0.89 |
ENSMUST00000137626.2
|
Mmp9
|
matrix metallopeptidase 9 |
| chr11_+_97340962 | 0.89 |
ENSMUST00000107601.8
|
Arhgap23
|
Rho GTPase activating protein 23 |
| chr11_-_78950641 | 0.87 |
ENSMUST00000226282.2
|
Ksr1
|
kinase suppressor of ras 1 |
| chr7_+_101060093 | 0.87 |
ENSMUST00000084894.15
|
Gm45837
|
predicted gene 45837 |
| chr16_+_87350202 | 0.87 |
ENSMUST00000026700.8
|
Map3k7cl
|
Map3k7 C-terminal like |
| chr2_+_122479770 | 0.86 |
ENSMUST00000047498.15
ENSMUST00000110512.4 |
AA467197
|
expressed sequence AA467197 |
| chr9_-_39515420 | 0.85 |
ENSMUST00000042485.11
ENSMUST00000141370.8 |
AW551984
|
expressed sequence AW551984 |
| chr7_-_28038129 | 0.85 |
ENSMUST00000209141.2
ENSMUST00000003527.10 |
Supt5
|
suppressor of Ty 5, DSIF elongation factor subunit |
| chr8_+_110595216 | 0.84 |
ENSMUST00000179721.8
ENSMUST00000034175.5 |
Phlpp2
|
PH domain and leucine rich repeat protein phosphatase 2 |
| chr6_-_68681962 | 0.84 |
ENSMUST00000103330.2
|
Igkv10-94
|
immunoglobulin kappa variable 10-94 |
| chr3_-_82053068 | 0.83 |
ENSMUST00000048976.8
|
Gucy1a1
|
guanylate cyclase 1, soluble, alpha 1 |
| chr10_+_122514669 | 0.83 |
ENSMUST00000161487.8
ENSMUST00000067918.12 |
Ppm1h
|
protein phosphatase 1H (PP2C domain containing) |
| chr9_-_66951114 | 0.82 |
ENSMUST00000113686.8
|
Tpm1
|
tropomyosin 1, alpha |
| chr11_+_59839032 | 0.81 |
ENSMUST00000081980.7
|
Med9
|
mediator complex subunit 9 |
| chr15_-_63869818 | 0.81 |
ENSMUST00000164532.3
|
Cyrib
|
CYFIP related Rac1 interactor B |
| chr6_-_69741999 | 0.80 |
ENSMUST00000103365.3
|
Igkv12-46
|
immunoglobulin kappa variable 12-46 |
| chr12_-_113733922 | 0.79 |
ENSMUST00000180013.3
|
Ighv2-9-1
|
immunoglobulin heavy variable 2-9-1 |
| chr12_+_17740831 | 0.77 |
ENSMUST00000071858.5
|
Hpcal1
|
hippocalcin-like 1 |
| chr3_-_116047148 | 0.77 |
ENSMUST00000090473.7
|
Gpr88
|
G-protein coupled receptor 88 |
| chr18_-_36648850 | 0.77 |
ENSMUST00000025363.7
|
Hbegf
|
heparin-binding EGF-like growth factor |
| chr6_+_67243967 | 0.75 |
ENSMUST00000203436.3
ENSMUST00000205106.3 ENSMUST00000204293.3 ENSMUST00000203077.3 ENSMUST00000204294.3 |
Serbp1
|
serpine1 mRNA binding protein 1 |
| chr6_-_124710030 | 0.74 |
ENSMUST00000173647.2
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr5_-_73413888 | 0.74 |
ENSMUST00000101127.12
|
Fryl
|
FRY like transcription coactivator |
| chr16_+_49620883 | 0.74 |
ENSMUST00000229640.2
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr14_-_30890544 | 0.74 |
ENSMUST00000036618.14
|
Stab1
|
stabilin 1 |
| chr9_-_62888156 | 0.73 |
ENSMUST00000098651.6
ENSMUST00000214830.2 |
Pias1
|
protein inhibitor of activated STAT 1 |
| chr9_-_27066685 | 0.73 |
ENSMUST00000034472.16
|
Jam3
|
junction adhesion molecule 3 |
| chr11_-_120538928 | 0.72 |
ENSMUST00000239158.2
ENSMUST00000026134.3 |
Myadml2
|
myeloid-associated differentiation marker-like 2 |
| chr7_+_75105282 | 0.72 |
ENSMUST00000207750.2
ENSMUST00000166315.7 |
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr11_+_96820220 | 0.72 |
ENSMUST00000062172.6
|
Prr15l
|
proline rich 15-like |
| chr17_-_43813664 | 0.71 |
ENSMUST00000024707.9
ENSMUST00000117137.8 |
Mep1a
|
meprin 1 alpha |
| chr3_+_96736600 | 0.70 |
ENSMUST00000135031.8
|
Pdzk1
|
PDZ domain containing 1 |
| chr11_+_99748741 | 0.70 |
ENSMUST00000107434.2
|
Gm11568
|
predicted gene 11568 |
| chr4_+_46489248 | 0.69 |
ENSMUST00000030018.5
|
Nans
|
N-acetylneuraminic acid synthase (sialic acid synthase) |
| chr5_-_134975773 | 0.69 |
ENSMUST00000051401.4
|
Cldn4
|
claudin 4 |
| chr17_+_28910714 | 0.69 |
ENSMUST00000233250.2
|
Mapk14
|
mitogen-activated protein kinase 14 |
| chr3_+_123240562 | 0.69 |
ENSMUST00000029603.10
|
Prss12
|
protease, serine 12 neurotrypsin (motopsin) |
| chr7_+_67602565 | 0.67 |
ENSMUST00000005671.10
|
Igf1r
|
insulin-like growth factor I receptor |
| chr19_+_10611574 | 0.66 |
ENSMUST00000055115.9
|
Vwce
|
von Willebrand factor C and EGF domains |
| chr9_-_66950991 | 0.66 |
ENSMUST00000113689.8
ENSMUST00000113684.8 |
Tpm1
|
tropomyosin 1, alpha |
| chr13_-_63006176 | 0.66 |
ENSMUST00000021907.9
|
Fbp2
|
fructose bisphosphatase 2 |
| chr6_-_124710084 | 0.65 |
ENSMUST00000112484.10
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr15_-_100322089 | 0.65 |
ENSMUST00000154331.2
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr4_-_94444975 | 0.65 |
ENSMUST00000030313.9
|
Caap1
|
caspase activity and apoptosis inhibitor 1 |
| chr12_-_40087393 | 0.65 |
ENSMUST00000146905.2
|
Arl4a
|
ADP-ribosylation factor-like 4A |
| chr10_-_7539009 | 0.64 |
ENSMUST00000163085.8
ENSMUST00000159917.8 |
Pcmt1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase 1 |
| chr9_+_57468217 | 0.63 |
ENSMUST00000045791.11
ENSMUST00000216986.2 |
Scamp2
|
secretory carrier membrane protein 2 |
| chr6_-_85351588 | 0.63 |
ENSMUST00000204087.2
|
Rab11fip5
|
RAB11 family interacting protein 5 (class I) |
| chr16_+_10652910 | 0.63 |
ENSMUST00000037913.9
|
Rmi2
|
RecQ mediated genome instability 2 |
| chr14_+_43951187 | 0.62 |
ENSMUST00000094051.6
|
Gm7324
|
predicted gene 7324 |
| chr19_-_41836514 | 0.62 |
ENSMUST00000059231.4
|
Frat2
|
frequently rearranged in advanced T cell lymphomas 2 |
| chr7_-_141579689 | 0.62 |
ENSMUST00000209732.2
|
Mob2
|
MOB kinase activator 2 |
| chr17_-_34174631 | 0.61 |
ENSMUST00000174609.9
ENSMUST00000008812.9 |
Rps18
|
ribosomal protein S18 |
| chr11_-_75329726 | 0.61 |
ENSMUST00000108437.8
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr2_-_180596469 | 0.60 |
ENSMUST00000148905.8
ENSMUST00000103053.10 ENSMUST00000108873.9 |
Nkain4
|
Na+/K+ transporting ATPase interacting 4 |
| chr5_+_64317550 | 0.60 |
ENSMUST00000101195.9
|
Tbc1d1
|
TBC1 domain family, member 1 |
| chrX_-_97934387 | 0.60 |
ENSMUST00000113826.8
ENSMUST00000033560.9 ENSMUST00000142267.2 |
Ophn1
|
oligophrenin 1 |
| chr9_-_62417780 | 0.60 |
ENSMUST00000164246.9
|
Coro2b
|
coronin, actin binding protein, 2B |
| chr10_-_7162196 | 0.60 |
ENSMUST00000015346.12
|
Cnksr3
|
Cnksr family member 3 |
| chr11_-_106679983 | 0.60 |
ENSMUST00000129585.8
|
Ddx5
|
DEAD box helicase 5 |
| chr13_-_76166789 | 0.59 |
ENSMUST00000179078.9
ENSMUST00000167271.9 |
Rfesd
|
Rieske (Fe-S) domain containing |
| chr13_+_24822619 | 0.59 |
ENSMUST00000110384.9
ENSMUST00000058009.16 ENSMUST00000038477.7 |
Ripor2
|
RHO family interacting cell polarization regulator 2 |
| chr2_+_32617671 | 0.59 |
ENSMUST00000113242.5
|
Sh2d3c
|
SH2 domain containing 3C |
| chr19_+_38384428 | 0.58 |
ENSMUST00000054098.4
|
Slc35g1
|
solute carrier family 35, member G1 |
| chr19_+_40600836 | 0.57 |
ENSMUST00000134063.8
|
Entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ascl2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 1.6 | 9.6 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 1.5 | 5.8 | GO:1900228 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 1.4 | 6.9 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 1.1 | 3.2 | GO:0002343 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 1.0 | 5.9 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.8 | 2.3 | GO:0019413 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 0.7 | 2.2 | GO:0002588 | myeloid dendritic cell activation involved in immune response(GO:0002277) positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.7 | 3.7 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.7 | 6.6 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.7 | 5.6 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.7 | 2.8 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.7 | 2.7 | GO:0045659 | negative regulation of neutrophil differentiation(GO:0045659) |
| 0.7 | 4.6 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.6 | 1.8 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.6 | 2.9 | GO:0007356 | thorax and anterior abdomen determination(GO:0007356) |
| 0.5 | 1.6 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.5 | 1.5 | GO:0046072 | dTDP biosynthetic process(GO:0006233) dTDP metabolic process(GO:0046072) |
| 0.5 | 2.0 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 0.5 | 2.8 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.5 | 11.3 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.4 | 1.7 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.4 | 1.7 | GO:1900756 | renal protein absorption(GO:0097017) protein processing in phagocytic vesicle(GO:1900756) positive regulation of keratinocyte apoptotic process(GO:1902174) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.4 | 2.6 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.4 | 1.1 | GO:0035638 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.3 | 1.4 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.3 | 2.7 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.3 | 1.9 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.3 | 0.6 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.3 | 2.5 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.3 | 0.9 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.3 | 1.4 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.3 | 7.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.3 | 3.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.3 | 0.3 | GO:0072166 | posterior mesonephric tubule development(GO:0072166) |
| 0.3 | 1.6 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.3 | 3.1 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.3 | 0.8 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 0.3 | 2.5 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.2 | 1.0 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.2 | 0.9 | GO:0070100 | regulation of chemokine-mediated signaling pathway(GO:0070099) negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.2 | 1.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.2 | 0.4 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.2 | 0.6 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) |
| 0.2 | 0.5 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.2 | 0.7 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.2 | 0.5 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.2 | 0.4 | GO:0042660 | positive regulation of cell fate specification(GO:0042660) |
| 0.2 | 1.4 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.2 | 0.5 | GO:1990773 | regulation of matrix metallopeptidase secretion(GO:1904464) matrix metallopeptidase secretion(GO:1990773) |
| 0.2 | 0.7 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.2 | 0.7 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.2 | 0.5 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.2 | 1.3 | GO:2001184 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.2 | 0.5 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 0.4 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.1 | 0.9 | GO:0015692 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 | 1.8 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.1 | 5.9 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 5.6 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.1 | 2.5 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 | 0.4 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 1.0 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.1 | 0.4 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 2.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.4 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.1 | 0.5 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 | 0.4 | GO:0009816 | defense response, incompatible interaction(GO:0009814) defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.1 | 0.8 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 1.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.8 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 3.5 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 1.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 1.0 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 1.9 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.5 | GO:0007494 | midgut development(GO:0007494) |
| 0.1 | 5.5 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.1 | 1.3 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.1 | 2.6 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.6 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.1 | 0.3 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 1.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.5 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 1.0 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.1 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.1 | 0.5 | GO:1903436 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.1 | 0.6 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 0.3 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 | 0.3 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 0.9 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 4.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.7 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.1 | 0.7 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.1 | 0.8 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 1.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.6 | GO:0003072 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.1 | 0.2 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.1 | 0.8 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 | 1.0 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.1 | 0.3 | GO:0090346 | regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.1 | 0.7 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 1.1 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.1 | 0.3 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.1 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 0.3 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.1 | 1.7 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 0.2 | GO:0090172 | attachment of spindle microtubules to kinetochore involved in homologous chromosome segregation(GO:0051455) microtubule cytoskeleton organization involved in homologous chromosome segregation(GO:0090172) homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.1 | 2.2 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.1 | 0.4 | GO:0021747 | synaptic vesicle targeting(GO:0016080) cochlear nucleus development(GO:0021747) |
| 0.1 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.2 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.3 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 1.1 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.1 | 1.1 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 0.2 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 0.2 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.7 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.4 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 0.0 | 0.4 | GO:0040016 | negative regulation of superoxide anion generation(GO:0032929) embryonic cleavage(GO:0040016) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:1902164 | positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.1 | GO:0010958 | regulation of amino acid import(GO:0010958) L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.0 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.0 | 0.3 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.2 | GO:0061590 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.2 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.3 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.8 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.9 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 1.6 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 0.4 | GO:0021995 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.0 | 0.1 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.0 | 0.5 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.3 | GO:0002568 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 5.1 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.5 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.2 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 1.0 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 1.6 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.8 | GO:0060416 | response to growth hormone(GO:0060416) |
| 0.0 | 0.8 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.4 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.0 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.9 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 1.6 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 2.8 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 1.1 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.2 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.7 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.3 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.5 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.4 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.3 | GO:0097284 | hepatocyte apoptotic process(GO:0097284) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.2 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.2 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.4 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) |
| 0.0 | 2.1 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.6 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.9 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 11.4 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 1.1 | 4.6 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 1.0 | 5.8 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.7 | 7.2 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.6 | 2.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.5 | 6.8 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.4 | 3.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.4 | 3.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.4 | 2.5 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.4 | 9.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.4 | 3.7 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.3 | 5.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.3 | 3.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.2 | 2.8 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 1.7 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 0.5 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.6 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 0.9 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 2.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.4 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.1 | 0.7 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 1.6 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.9 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 9.8 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 0.3 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.1 | 0.4 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 1.0 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 5.9 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.3 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 0.4 | GO:0000938 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.1 | 0.7 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 2.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.4 | GO:0071547 | piP-body(GO:0071547) |
| 0.1 | 0.4 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 2.6 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 1.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.9 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.5 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 0.4 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 1.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 8.6 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 1.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.5 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.8 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 1.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.4 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 1.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 1.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 2.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 6.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 3.1 | GO:0019814 | immunoglobulin complex(GO:0019814) immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 2.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 3.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 1.6 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 2.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 3.1 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 2.5 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 3.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.9 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.6 | 1.8 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.6 | 2.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.5 | 2.5 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.5 | 1.5 | GO:0004798 | thymidylate kinase activity(GO:0004798) |
| 0.5 | 11.4 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.5 | 2.3 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.4 | 3.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.4 | 1.2 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 0.4 | 0.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.3 | 3.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.3 | 1.0 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.3 | 9.6 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.3 | 3.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.3 | 3.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 1.8 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 2.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.2 | 1.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.2 | 1.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 3.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.2 | 5.8 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.2 | 0.5 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.2 | 10.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.2 | 7.0 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.2 | 1.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.9 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 3.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.6 | GO:0051435 | BH4 domain binding(GO:0051435) |
| 0.1 | 0.7 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 5.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.0 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 1.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.5 | GO:0038100 | nodal binding(GO:0038100) |
| 0.1 | 0.5 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 0.9 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 8.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 4.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.0 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 | 1.4 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 8.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.7 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 1.3 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 5.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.3 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 1.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.1 | 1.7 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.3 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.1 | 0.5 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 2.6 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.7 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.1 | 0.8 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.3 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 2.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 9.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.3 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.1 | 0.7 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.7 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.5 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.6 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.3 | GO:0008941 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.1 | 0.7 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 1.1 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.1 | 0.6 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 0.2 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.1 | 0.9 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 2.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.2 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 0.2 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.1 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.4 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.5 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 6.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.7 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.3 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.7 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 7.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.5 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 1.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 1.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 1.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.3 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 1.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.5 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.5 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 12.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0002135 | CTP binding(GO:0002135) |
| 0.0 | 0.1 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.0 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.4 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 1.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 1.1 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 7.3 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.2 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.7 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 9.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 1.8 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.2 | 8.8 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 7.7 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 2.5 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 3.1 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 2.6 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 4.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 3.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 5.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 1.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 0.7 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 3.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 2.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 5.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 1.2 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 1.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.6 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.0 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.9 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.9 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 1.2 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.2 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 2.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.2 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 3.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | PID BCR 5PATHWAY | BCR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.3 | 7.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.3 | 10.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.3 | 12.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.3 | 6.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 3.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.2 | 2.7 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.2 | 5.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 12.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 3.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 7.9 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.2 | 2.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 5.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 5.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 2.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 4.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 2.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 0.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 0.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 3.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 4.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.7 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 3.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 1.3 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 6.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.3 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.3 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |