Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Atf2_Ikzf1
Z-value: 0.71
Transcription factors associated with Atf2_Ikzf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
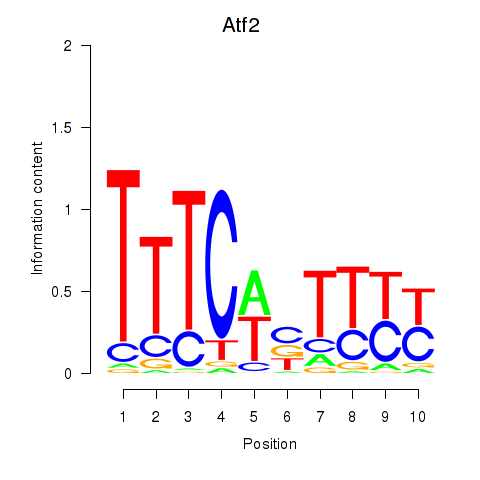
Atf2
|
ENSMUSG00000027104.19 | activating transcription factor 2 |
|
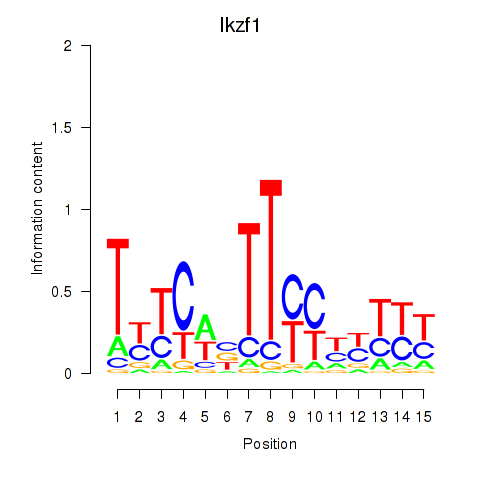
Ikzf1
|
ENSMUSG00000018654.18 | IKAROS family zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ikzf1 | mm39_v1_chr11_+_11634967_11635027 | -0.73 | 3.7e-07 | Click! |
| Atf2 | mm39_v1_chr2_-_73722932_73722983 | -0.49 | 2.3e-03 | Click! |
Activity profile of Atf2_Ikzf1 motif
Sorted Z-values of Atf2_Ikzf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_55152002 | 4.79 |
ENSMUST00000003569.6
|
Inmt
|
indolethylamine N-methyltransferase |
| chr19_+_40078132 | 3.38 |
ENSMUST00000068094.13
ENSMUST00000080171.3 |
Cyp2c50
|
cytochrome P450, family 2, subfamily c, polypeptide 50 |
| chr7_+_26534730 | 3.15 |
ENSMUST00000005685.15
|
Cyp2a5
|
cytochrome P450, family 2, subfamily a, polypeptide 5 |
| chr19_-_40062174 | 3.05 |
ENSMUST00000048959.5
|
Cyp2c54
|
cytochrome P450, family 2, subfamily c, polypeptide 54 |
| chr3_+_97536120 | 2.53 |
ENSMUST00000107050.8
ENSMUST00000029729.15 ENSMUST00000107049.2 |
Fmo5
|
flavin containing monooxygenase 5 |
| chr19_+_39980868 | 2.50 |
ENSMUST00000049178.3
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr6_-_114898739 | 1.93 |
ENSMUST00000032459.14
|
Vgll4
|
vestigial like family member 4 |
| chr19_-_20704896 | 1.78 |
ENSMUST00000025656.4
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7 |
| chr6_-_124519240 | 1.71 |
ENSMUST00000159463.8
ENSMUST00000162844.2 ENSMUST00000160505.8 ENSMUST00000162443.8 |
C1s1
|
complement component 1, s subcomponent 1 |
| chr4_-_60457902 | 1.59 |
ENSMUST00000084548.11
ENSMUST00000103012.10 ENSMUST00000107499.4 |
Mup1
|
major urinary protein 1 |
| chr17_+_35481702 | 1.59 |
ENSMUST00000172785.8
|
H2-D1
|
histocompatibility 2, D region locus 1 |
| chr8_-_85500010 | 1.53 |
ENSMUST00000109764.8
|
Nfix
|
nuclear factor I/X |
| chr15_+_100202079 | 1.52 |
ENSMUST00000230252.2
ENSMUST00000231166.2 |
Mettl7a1
|
methyltransferase like 7A1 |
| chr15_+_100202061 | 1.51 |
ENSMUST00000229574.2
ENSMUST00000229217.2 |
Mettl7a1
|
methyltransferase like 7A1 |
| chr2_-_173060647 | 1.51 |
ENSMUST00000109116.3
ENSMUST00000029018.14 |
Zbp1
|
Z-DNA binding protein 1 |
| chr5_+_115061293 | 1.49 |
ENSMUST00000031540.11
ENSMUST00000112143.4 |
Oasl1
|
2'-5' oligoadenylate synthetase-like 1 |
| chr9_-_106353792 | 1.49 |
ENSMUST00000214682.2
ENSMUST00000112479.9 |
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr7_+_101027390 | 1.48 |
ENSMUST00000084895.12
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr6_-_23839136 | 1.46 |
ENSMUST00000166458.9
ENSMUST00000142913.9 ENSMUST00000069074.14 ENSMUST00000115361.9 ENSMUST00000018122.14 ENSMUST00000115356.3 |
Cadps2
|
Ca2+-dependent activator protein for secretion 2 |
| chr7_-_48493388 | 1.43 |
ENSMUST00000167786.4
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr6_+_121320008 | 1.42 |
ENSMUST00000166457.8
|
Slc6a12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 |
| chr6_+_125298372 | 1.42 |
ENSMUST00000176442.8
ENSMUST00000177329.2 |
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr15_+_10314173 | 1.40 |
ENSMUST00000127467.3
|
Prlr
|
prolactin receptor |
| chr10_+_29019645 | 1.39 |
ENSMUST00000092629.4
|
Soga3
|
SOGA family member 3 |
| chr17_-_34219225 | 1.39 |
ENSMUST00000238098.2
ENSMUST00000087189.7 ENSMUST00000173075.3 ENSMUST00000172912.8 ENSMUST00000236740.2 ENSMUST00000025181.18 |
H2-K1
|
histocompatibility 2, K1, K region |
| chr15_+_100202021 | 1.37 |
ENSMUST00000230472.2
|
Mettl7a1
|
methyltransferase like 7A1 |
| chr14_+_55798517 | 1.35 |
ENSMUST00000117701.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr12_-_84497718 | 1.32 |
ENSMUST00000085192.7
ENSMUST00000220491.2 |
Aldh6a1
|
aldehyde dehydrogenase family 6, subfamily A1 |
| chr10_+_116111441 | 1.31 |
ENSMUST00000218553.2
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr8_-_25066313 | 1.28 |
ENSMUST00000121992.2
|
Ido2
|
indoleamine 2,3-dioxygenase 2 |
| chr6_+_41498716 | 1.27 |
ENSMUST00000070380.5
|
Prss2
|
protease, serine 2 |
| chr1_-_192946359 | 1.23 |
ENSMUST00000161737.8
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr17_-_34218301 | 1.22 |
ENSMUST00000235463.2
|
H2-K1
|
histocompatibility 2, K1, K region |
| chr6_+_43242516 | 1.22 |
ENSMUST00000031750.14
|
Arhgef5
|
Rho guanine nucleotide exchange factor (GEF) 5 |
| chr1_-_169575203 | 1.21 |
ENSMUST00000027991.12
ENSMUST00000111357.2 |
Rgs4
|
regulator of G-protein signaling 4 |
| chr9_-_106353571 | 1.21 |
ENSMUST00000123555.8
ENSMUST00000125850.2 |
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr2_+_58645189 | 1.18 |
ENSMUST00000102755.4
ENSMUST00000230627.2 ENSMUST00000229923.2 |
Upp2
|
uridine phosphorylase 2 |
| chr1_+_167445815 | 1.15 |
ENSMUST00000111380.2
|
Rxrg
|
retinoid X receptor gamma |
| chr10_-_43880353 | 1.15 |
ENSMUST00000020017.14
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr3_+_20039775 | 1.14 |
ENSMUST00000172860.2
|
Cp
|
ceruloplasmin |
| chr11_-_116080361 | 1.14 |
ENSMUST00000148601.2
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr11_+_68858942 | 1.10 |
ENSMUST00000102606.10
ENSMUST00000018884.6 |
Slc25a35
|
solute carrier family 25, member 35 |
| chr2_+_43445333 | 1.08 |
ENSMUST00000028223.9
ENSMUST00000112826.8 |
Kynu
|
kynureninase |
| chr6_+_121320339 | 1.08 |
ENSMUST00000168295.2
|
Slc6a12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 |
| chr5_+_87148697 | 1.07 |
ENSMUST00000031186.9
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chrX_+_106299484 | 1.06 |
ENSMUST00000101294.9
ENSMUST00000118820.8 ENSMUST00000120971.8 |
Gpr174
|
G protein-coupled receptor 174 |
| chr7_-_126184935 | 1.06 |
ENSMUST00000084589.11
|
Cln3
|
ceroid lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) |
| chr1_+_155911518 | 1.04 |
ENSMUST00000133152.2
|
Tor1aip2
|
torsin A interacting protein 2 |
| chr15_+_100202642 | 1.04 |
ENSMUST00000067752.5
ENSMUST00000229588.2 |
Mettl7a1
|
methyltransferase like 7A1 |
| chr11_+_60668350 | 1.03 |
ENSMUST00000102667.5
ENSMUST00000056907.7 |
Smcr8
|
Smith-Magenis syndrome chromosome region, candidate 8 homolog (human) |
| chr9_-_91247809 | 1.02 |
ENSMUST00000034927.13
|
Zic1
|
zinc finger protein of the cerebellum 1 |
| chr9_-_50466470 | 1.02 |
ENSMUST00000119103.2
|
Bco2
|
beta-carotene oxygenase 2 |
| chr2_+_58644922 | 1.01 |
ENSMUST00000059102.13
|
Upp2
|
uridine phosphorylase 2 |
| chr5_-_87682972 | 1.00 |
ENSMUST00000120150.2
|
Sult1b1
|
sulfotransferase family 1B, member 1 |
| chr19_+_34560922 | 1.00 |
ENSMUST00000102825.4
|
Ifit3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr7_-_141015240 | 0.98 |
ENSMUST00000138865.8
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr7_-_80051455 | 0.98 |
ENSMUST00000120753.3
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr12_+_116239006 | 0.97 |
ENSMUST00000090195.5
|
Gm11027
|
predicted gene 11027 |
| chr6_-_136852792 | 0.97 |
ENSMUST00000032342.3
|
Mgp
|
matrix Gla protein |
| chr1_-_180021218 | 0.96 |
ENSMUST00000159914.8
|
Coq8a
|
coenzyme Q8A |
| chr10_+_87694924 | 0.96 |
ENSMUST00000095360.11
|
Igf1
|
insulin-like growth factor 1 |
| chr19_-_43879031 | 0.96 |
ENSMUST00000212048.2
|
Dnmbp
|
dynamin binding protein |
| chr5_-_121710768 | 0.95 |
ENSMUST00000200541.5
|
Aldh2
|
aldehyde dehydrogenase 2, mitochondrial |
| chr1_-_192923816 | 0.95 |
ENSMUST00000160929.8
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr15_+_32920869 | 0.95 |
ENSMUST00000022871.7
|
Sdc2
|
syndecan 2 |
| chr4_-_96552349 | 0.93 |
ENSMUST00000030299.8
|
Cyp2j5
|
cytochrome P450, family 2, subfamily j, polypeptide 5 |
| chr3_+_20011251 | 0.93 |
ENSMUST00000108328.8
|
Cp
|
ceruloplasmin |
| chr2_+_43445359 | 0.92 |
ENSMUST00000050511.7
|
Kynu
|
kynureninase |
| chr5_-_87572060 | 0.91 |
ENSMUST00000072818.6
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr3_+_95190255 | 0.91 |
ENSMUST00000039537.14
ENSMUST00000107187.9 |
Mindy1
|
MINDY lysine 48 deubiquitinase 1 |
| chr17_-_57529827 | 0.90 |
ENSMUST00000177425.2
|
C3
|
complement component 3 |
| chr11_+_75358866 | 0.90 |
ENSMUST00000043598.14
ENSMUST00000108435.2 |
Tlcd2
|
TLC domain containing 2 |
| chr7_+_26006594 | 0.89 |
ENSMUST00000098657.5
|
Cyp2a4
|
cytochrome P450, family 2, subfamily a, polypeptide 4 |
| chr4_+_144619397 | 0.89 |
ENSMUST00000105744.8
ENSMUST00000171001.8 |
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr17_+_35658131 | 0.88 |
ENSMUST00000071951.14
ENSMUST00000116598.10 ENSMUST00000078205.14 ENSMUST00000076256.8 |
H2-Q7
|
histocompatibility 2, Q region locus 7 |
| chr18_+_24786748 | 0.88 |
ENSMUST00000068006.9
|
Mocos
|
molybdenum cofactor sulfurase |
| chr19_+_3372296 | 0.85 |
ENSMUST00000237938.2
|
Cpt1a
|
carnitine palmitoyltransferase 1a, liver |
| chr3_+_59989282 | 0.84 |
ENSMUST00000029326.6
|
Sucnr1
|
succinate receptor 1 |
| chr16_+_43067641 | 0.84 |
ENSMUST00000079441.13
ENSMUST00000114691.8 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr16_+_23338960 | 0.82 |
ENSMUST00000211460.2
ENSMUST00000210658.2 ENSMUST00000209198.2 ENSMUST00000210371.2 ENSMUST00000211499.2 ENSMUST00000210795.2 ENSMUST00000209422.2 |
Gm45338
Rtp4
|
predicted gene 45338 receptor transporter protein 4 |
| chr13_+_42862957 | 0.82 |
ENSMUST00000066928.12
ENSMUST00000148891.8 |
Phactr1
|
phosphatase and actin regulator 1 |
| chr2_-_35994819 | 0.81 |
ENSMUST00000148852.4
|
Lhx6
|
LIM homeobox protein 6 |
| chr8_-_85500998 | 0.80 |
ENSMUST00000109762.8
|
Nfix
|
nuclear factor I/X |
| chr9_-_117080869 | 0.79 |
ENSMUST00000172564.3
|
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chr13_-_78347876 | 0.77 |
ENSMUST00000091458.13
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr16_-_45975440 | 0.76 |
ENSMUST00000059524.7
|
Gm4737
|
predicted gene 4737 |
| chr5_+_90666791 | 0.75 |
ENSMUST00000113179.9
ENSMUST00000128740.2 |
Afm
|
afamin |
| chr5_-_89605622 | 0.75 |
ENSMUST00000049209.13
|
Gc
|
vitamin D binding protein |
| chr1_+_155911451 | 0.74 |
ENSMUST00000111754.9
|
Tor1aip2
|
torsin A interacting protein 2 |
| chr3_+_20011405 | 0.74 |
ENSMUST00000108325.9
|
Cp
|
ceruloplasmin |
| chr19_+_38995463 | 0.74 |
ENSMUST00000025966.5
|
Cyp2c55
|
cytochrome P450, family 2, subfamily c, polypeptide 55 |
| chr11_+_66915969 | 0.74 |
ENSMUST00000079077.12
ENSMUST00000061786.6 |
Tmem220
|
transmembrane protein 220 |
| chr17_-_84154173 | 0.74 |
ENSMUST00000000687.9
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr17_+_35780977 | 0.73 |
ENSMUST00000174525.8
ENSMUST00000068291.7 |
H2-Q10
|
histocompatibility 2, Q region locus 10 |
| chr4_-_49549489 | 0.73 |
ENSMUST00000029987.10
|
Aldob
|
aldolase B, fructose-bisphosphate |
| chr4_-_62069046 | 0.72 |
ENSMUST00000077719.4
|
Mup21
|
major urinary protein 21 |
| chr3_+_20011201 | 0.72 |
ENSMUST00000091309.12
ENSMUST00000108329.8 ENSMUST00000003714.13 |
Cp
|
ceruloplasmin |
| chr18_-_75094323 | 0.71 |
ENSMUST00000066532.5
|
Lipg
|
lipase, endothelial |
| chr2_-_164699462 | 0.71 |
ENSMUST00000109316.8
ENSMUST00000156255.8 ENSMUST00000128110.2 ENSMUST00000109317.10 ENSMUST00000059954.14 |
Pltp
|
phospholipid transfer protein |
| chr2_-_32851586 | 0.70 |
ENSMUST00000133832.8
ENSMUST00000124492.8 ENSMUST00000145578.8 ENSMUST00000127321.8 ENSMUST00000113200.8 |
Lrsam1
|
leucine rich repeat and sterile alpha motif containing 1 |
| chr17_-_56312555 | 0.70 |
ENSMUST00000043785.8
|
Stap2
|
signal transducing adaptor family member 2 |
| chr7_-_30672747 | 0.70 |
ENSMUST00000205961.2
|
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr3_+_94280101 | 0.70 |
ENSMUST00000029795.10
|
Rorc
|
RAR-related orphan receptor gamma |
| chr7_+_107166653 | 0.70 |
ENSMUST00000120990.2
|
Olfml1
|
olfactomedin-like 1 |
| chr17_+_12597490 | 0.70 |
ENSMUST00000014578.7
|
Plg
|
plasminogen |
| chr9_+_90045219 | 0.69 |
ENSMUST00000147250.8
ENSMUST00000113060.3 |
Adamts7
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 7 |
| chr9_+_90045109 | 0.68 |
ENSMUST00000113059.8
ENSMUST00000167122.8 |
Adamts7
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 7 |
| chr6_+_121277693 | 0.68 |
ENSMUST00000142419.2
|
Slc6a13
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 13 |
| chr5_+_90608751 | 0.68 |
ENSMUST00000031314.10
|
Alb
|
albumin |
| chr1_-_133849131 | 0.68 |
ENSMUST00000048432.6
|
Prelp
|
proline arginine-rich end leucine-rich repeat |
| chr6_+_34723304 | 0.67 |
ENSMUST00000142716.3
|
Cald1
|
caldesmon 1 |
| chr7_+_99655611 | 0.67 |
ENSMUST00000032977.11
|
Chrdl2
|
chordin-like 2 |
| chr9_-_91247831 | 0.66 |
ENSMUST00000065360.5
|
Zic1
|
zinc finger protein of the cerebellum 1 |
| chr6_-_93889483 | 0.66 |
ENSMUST00000205116.3
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr17_-_84154196 | 0.65 |
ENSMUST00000234214.2
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr2_+_67578556 | 0.65 |
ENSMUST00000180887.2
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr10_-_127206300 | 0.64 |
ENSMUST00000026472.10
|
Inhbc
|
inhibin beta-C |
| chr3_+_99161070 | 0.63 |
ENSMUST00000029462.10
|
Tbx15
|
T-box 15 |
| chr16_+_43184191 | 0.63 |
ENSMUST00000156367.8
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr1_-_170803680 | 0.63 |
ENSMUST00000027966.14
ENSMUST00000081103.12 ENSMUST00000159688.2 |
Fcgr2b
|
Fc receptor, IgG, low affinity IIb |
| chr16_-_23339329 | 0.63 |
ENSMUST00000230040.2
ENSMUST00000229619.2 |
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr6_+_121323577 | 0.63 |
ENSMUST00000032200.16
|
Slc6a12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 |
| chr8_-_71990085 | 0.62 |
ENSMUST00000051672.9
|
Bst2
|
bone marrow stromal cell antigen 2 |
| chr17_+_85335775 | 0.61 |
ENSMUST00000024944.9
|
Slc3a1
|
solute carrier family 3, member 1 |
| chr19_-_58443593 | 0.61 |
ENSMUST00000135730.2
ENSMUST00000152507.8 |
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr1_-_155022501 | 0.61 |
ENSMUST00000027744.10
|
Mr1
|
major histocompatibility complex, class I-related |
| chr7_+_51528715 | 0.60 |
ENSMUST00000051912.13
|
Gas2
|
growth arrest specific 2 |
| chr12_-_90705212 | 0.59 |
ENSMUST00000082432.6
|
Dio2
|
deiodinase, iodothyronine, type II |
| chr17_+_56312672 | 0.58 |
ENSMUST00000133998.8
|
Mpnd
|
MPN domain containing |
| chr14_-_80008745 | 0.58 |
ENSMUST00000039568.11
ENSMUST00000195355.2 |
Pcdh8
|
protocadherin 8 |
| chr7_+_107166925 | 0.58 |
ENSMUST00000239087.2
|
Olfml1
|
olfactomedin-like 1 |
| chr17_-_47063095 | 0.58 |
ENSMUST00000121671.2
ENSMUST00000059844.13 |
Cnpy3
|
canopy FGF signaling regulator 3 |
| chr13_+_41071077 | 0.58 |
ENSMUST00000067778.8
ENSMUST00000225759.2 |
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr6_+_41279199 | 0.57 |
ENSMUST00000031913.5
|
Try4
|
trypsin 4 |
| chr1_-_46927230 | 0.57 |
ENSMUST00000185520.2
|
Slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr15_+_10216041 | 0.56 |
ENSMUST00000130720.8
|
Prlr
|
prolactin receptor |
| chr14_-_64654397 | 0.56 |
ENSMUST00000210428.2
|
Msra
|
methionine sulfoxide reductase A |
| chr14_-_64654592 | 0.56 |
ENSMUST00000210363.2
|
Msra
|
methionine sulfoxide reductase A |
| chr19_+_53891730 | 0.56 |
ENSMUST00000025931.14
|
Pdcd4
|
programmed cell death 4 |
| chr7_-_97228589 | 0.56 |
ENSMUST00000151840.2
ENSMUST00000135998.8 ENSMUST00000144858.8 ENSMUST00000146605.8 ENSMUST00000072725.12 ENSMUST00000138060.3 ENSMUST00000154853.8 ENSMUST00000136757.8 ENSMUST00000124552.3 |
Aamdc
|
adipogenesis associated Mth938 domain containing |
| chr9_+_74883377 | 0.56 |
ENSMUST00000081746.7
|
Fam214a
|
family with sequence similarity 214, member A |
| chr11_-_21320452 | 0.55 |
ENSMUST00000102875.11
|
Ugp2
|
UDP-glucose pyrophosphorylase 2 |
| chr11_-_120464062 | 0.55 |
ENSMUST00000026122.11
|
P4hb
|
prolyl 4-hydroxylase, beta polypeptide |
| chr4_+_138694422 | 0.55 |
ENSMUST00000116094.5
ENSMUST00000239443.2 |
Rnf186
|
ring finger protein 186 |
| chrX_+_149981074 | 0.55 |
ENSMUST00000184730.8
ENSMUST00000184392.8 ENSMUST00000096285.5 |
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr19_+_53891802 | 0.54 |
ENSMUST00000165617.3
|
Pdcd4
|
programmed cell death 4 |
| chr18_+_56565188 | 0.54 |
ENSMUST00000070166.6
|
Gramd3
|
GRAM domain containing 3 |
| chr2_-_32314017 | 0.54 |
ENSMUST00000113307.9
|
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr10_+_122284404 | 0.54 |
ENSMUST00000020323.7
|
Avpr1a
|
arginine vasopressin receptor 1A |
| chr4_-_43656437 | 0.54 |
ENSMUST00000030192.5
|
Hint2
|
histidine triad nucleotide binding protein 2 |
| chr4_+_144619647 | 0.53 |
ENSMUST00000154208.8
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr15_-_5093222 | 0.53 |
ENSMUST00000110689.5
|
C7
|
complement component 7 |
| chr7_+_51528788 | 0.53 |
ENSMUST00000107591.9
|
Gas2
|
growth arrest specific 2 |
| chr17_-_34822649 | 0.52 |
ENSMUST00000015622.8
|
Rnf5
|
ring finger protein 5 |
| chr8_-_41668182 | 0.52 |
ENSMUST00000034003.5
|
Fgl1
|
fibrinogen-like protein 1 |
| chrX_+_72830668 | 0.52 |
ENSMUST00000002090.3
|
Ssr4
|
signal sequence receptor, delta |
| chr2_-_180798785 | 0.52 |
ENSMUST00000055990.8
|
Eef1a2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr4_+_144619696 | 0.52 |
ENSMUST00000142808.8
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr6_-_122317156 | 0.51 |
ENSMUST00000159384.8
|
Phc1
|
polyhomeotic 1 |
| chr9_+_44290832 | 0.51 |
ENSMUST00000161318.8
ENSMUST00000217019.2 ENSMUST00000160902.8 |
Hyou1
|
hypoxia up-regulated 1 |
| chr7_-_30754240 | 0.51 |
ENSMUST00000206860.2
ENSMUST00000071697.11 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr7_-_30754223 | 0.51 |
ENSMUST00000206012.2
ENSMUST00000108110.5 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr11_+_69983459 | 0.51 |
ENSMUST00000102572.8
|
Asgr2
|
asialoglycoprotein receptor 2 |
| chr14_+_50595361 | 0.50 |
ENSMUST00000185091.2
|
Tlr11
|
toll-like receptor 11 |
| chr1_+_140173787 | 0.50 |
ENSMUST00000239229.2
ENSMUST00000120709.8 ENSMUST00000120796.8 ENSMUST00000119786.8 |
Kcnt2
|
potassium channel, subfamily T, member 2 |
| chr14_-_124914516 | 0.50 |
ENSMUST00000095529.10
|
Fgf14
|
fibroblast growth factor 14 |
| chrX_+_72830607 | 0.50 |
ENSMUST00000166518.8
|
Ssr4
|
signal sequence receptor, delta |
| chr7_-_30754193 | 0.50 |
ENSMUST00000205778.2
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr8_-_111532495 | 0.49 |
ENSMUST00000150680.2
ENSMUST00000076846.11 |
Il34
|
interleukin 34 |
| chr7_+_130537902 | 0.49 |
ENSMUST00000006367.8
|
Htra1
|
HtrA serine peptidase 1 |
| chr11_+_43046476 | 0.49 |
ENSMUST00000238415.2
|
Atp10b
|
ATPase, class V, type 10B |
| chr17_-_36207965 | 0.49 |
ENSMUST00000150056.2
ENSMUST00000156817.2 ENSMUST00000146451.8 ENSMUST00000148482.8 |
2310061I04Rik
|
RIKEN cDNA 2310061I04 gene |
| chr1_-_65218217 | 0.49 |
ENSMUST00000097709.11
|
Idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr14_+_70314652 | 0.49 |
ENSMUST00000035908.3
|
Egr3
|
early growth response 3 |
| chr3_+_142300601 | 0.48 |
ENSMUST00000029936.5
|
Gbp2b
|
guanylate binding protein 2b |
| chr1_+_158189831 | 0.48 |
ENSMUST00000193042.6
ENSMUST00000046110.16 |
Astn1
|
astrotactin 1 |
| chr9_-_21838758 | 0.48 |
ENSMUST00000046831.11
ENSMUST00000238930.2 |
Tmem205
|
transmembrane protein 205 |
| chr1_-_66984521 | 0.48 |
ENSMUST00000160100.2
|
Myl1
|
myosin, light polypeptide 1 |
| chr14_-_67106037 | 0.48 |
ENSMUST00000022629.9
|
Dpysl2
|
dihydropyrimidinase-like 2 |
| chr17_+_34138699 | 0.48 |
ENSMUST00000234320.2
|
Tapbp
|
TAP binding protein |
| chr17_+_34482183 | 0.48 |
ENSMUST00000040828.7
ENSMUST00000237342.2 ENSMUST00000237866.2 |
H2-Ab1
|
histocompatibility 2, class II antigen A, beta 1 |
| chr18_-_60860594 | 0.48 |
ENSMUST00000235795.2
|
Ndst1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr8_+_129085719 | 0.48 |
ENSMUST00000026917.10
|
Nrp1
|
neuropilin 1 |
| chr6_+_121277186 | 0.48 |
ENSMUST00000064580.14
|
Slc6a13
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 13 |
| chr9_+_119978773 | 0.48 |
ENSMUST00000068698.15
ENSMUST00000215512.2 ENSMUST00000111627.3 ENSMUST00000093773.8 |
Mobp
|
myelin-associated oligodendrocytic basic protein |
| chr17_+_34138611 | 0.47 |
ENSMUST00000234247.2
|
Tapbp
|
TAP binding protein |
| chr1_+_42734889 | 0.47 |
ENSMUST00000054883.4
|
Pou3f3
|
POU domain, class 3, transcription factor 3 |
| chr11_-_120463667 | 0.47 |
ENSMUST00000168360.2
|
P4hb
|
prolyl 4-hydroxylase, beta polypeptide |
| chr4_-_84593226 | 0.47 |
ENSMUST00000175800.8
ENSMUST00000176418.8 ENSMUST00000175969.8 ENSMUST00000176370.2 ENSMUST00000176947.8 ENSMUST00000102820.9 ENSMUST00000107198.9 ENSMUST00000175756.8 ENSMUST00000176691.8 |
Bnc2
|
basonuclin 2 |
| chr7_+_43361930 | 0.47 |
ENSMUST00000066834.8
|
Klk13
|
kallikrein related-peptidase 13 |
| chr11_+_86375441 | 0.47 |
ENSMUST00000020827.7
|
Rnft1
|
ring finger protein, transmembrane 1 |
| chr3_+_96484294 | 0.47 |
ENSMUST00000148290.2
|
Gm16253
|
predicted gene 16253 |
| chr11_+_69983531 | 0.47 |
ENSMUST00000124721.2
|
Asgr2
|
asialoglycoprotein receptor 2 |
| chr16_+_42727926 | 0.46 |
ENSMUST00000151244.8
ENSMUST00000114694.9 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr11_+_69983479 | 0.46 |
ENSMUST00000143772.8
|
Asgr2
|
asialoglycoprotein receptor 2 |
| chr1_-_66984178 | 0.46 |
ENSMUST00000027151.12
|
Myl1
|
myosin, light polypeptide 1 |
| chr12_-_80159768 | 0.46 |
ENSMUST00000219642.2
ENSMUST00000165114.2 ENSMUST00000218835.2 ENSMUST00000021552.3 |
Zfp36l1
|
zinc finger protein 36, C3H type-like 1 |
| chr11_-_53313950 | 0.45 |
ENSMUST00000036045.6
|
Leap2
|
liver-expressed antimicrobial peptide 2 |
| chr1_-_170804116 | 0.45 |
ENSMUST00000159969.8
|
Fcgr2b
|
Fc receptor, IgG, low affinity IIb |
| chr6_-_28134544 | 0.45 |
ENSMUST00000115323.8
|
Grm8
|
glutamate receptor, metabotropic 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Atf2_Ikzf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.2 | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.7 | 2.2 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.7 | 2.0 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.6 | 3.1 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.6 | 7.2 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.5 | 3.1 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.5 | 1.4 | GO:1903920 | positive regulation of actin filament severing(GO:1903920) |
| 0.4 | 1.1 | GO:1904761 | cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) negative regulation of myofibroblast differentiation(GO:1904761) negative regulation of vascular smooth muscle cell differentiation(GO:1905064) |
| 0.3 | 1.3 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.3 | 2.0 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.3 | 1.2 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.3 | 0.9 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) |
| 0.3 | 1.1 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) negative regulation of hypersensitivity(GO:0002884) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.2 | 1.0 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.2 | 0.7 | GO:0060067 | cervix development(GO:0060067) |
| 0.2 | 1.0 | GO:0050822 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.2 | 0.7 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.2 | 0.9 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.2 | 0.7 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.2 | 1.4 | GO:0042436 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.2 | 0.7 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.2 | 0.6 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.2 | 1.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.2 | 6.5 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.2 | 1.4 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.2 | 1.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.2 | 1.1 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.2 | 0.6 | GO:0019255 | UDP-glucose metabolic process(GO:0006011) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.2 | 1.5 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.2 | 0.5 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.2 | 0.5 | GO:0021627 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.2 | 0.5 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.2 | 0.5 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 0.2 | 2.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 0.7 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.2 | 0.5 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.2 | 1.2 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.2 | 0.6 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 | 0.5 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.1 | 0.4 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.1 | 0.1 | GO:0060197 | cloacal septation(GO:0060197) |
| 0.1 | 0.7 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 2.6 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.4 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 1.0 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.1 | 2.8 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.4 | GO:0003167 | atrioventricular bundle cell differentiation(GO:0003167) |
| 0.1 | 0.4 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) olfactory pit development(GO:0060166) |
| 0.1 | 0.6 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.1 | 0.5 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.5 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.1 | 2.8 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.1 | 0.7 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.1 | 0.5 | GO:0072233 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.1 | 0.4 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 0.7 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 1.0 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 1.0 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 1.3 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 0.9 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.1 | 0.2 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.1 | 0.4 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.1 | 0.6 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.1 | 0.8 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.1 | 0.2 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.1 | 0.4 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.3 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.1 | 0.1 | GO:0035566 | regulation of metanephros size(GO:0035566) |
| 0.1 | 0.2 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.1 | 0.5 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.1 | 0.5 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 1.1 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.1 | 0.3 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.1 | 1.0 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 1.8 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 0.6 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 1.6 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.1 | 0.1 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.1 | 0.3 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.1 | 0.3 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.1 | 0.4 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.3 | GO:1904093 | regulation of autophagic cell death(GO:1904092) negative regulation of autophagic cell death(GO:1904093) |
| 0.1 | 3.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.3 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.1 | 0.7 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.3 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.1 | 0.3 | GO:0097402 | neuroblast migration(GO:0097402) |
| 0.1 | 0.2 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 0.3 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 | 0.3 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.1 | 0.5 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.3 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 0.3 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.2 | GO:1900239 | regulation of phenotypic switching(GO:1900239) |
| 0.1 | 0.2 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.1 | 0.5 | GO:2000054 | regulation of mismatch repair(GO:0032423) regulation of chondrocyte development(GO:0061181) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 0.2 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.1 | 0.2 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 0.1 | 0.3 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.1 | 2.0 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 0.2 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 0.1 | GO:0046101 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.1 | 0.3 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 0.3 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.1 | 2.8 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 0.1 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.2 | GO:0048936 | visceral motor neuron differentiation(GO:0021524) peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.1 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.1 | 0.4 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.1 | 0.4 | GO:0048105 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.1 | 1.3 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.1 | 0.3 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.1 | GO:0003133 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.1 | 0.7 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.1 | 0.1 | GO:0072198 | mesenchymal cell proliferation involved in ureter development(GO:0072198) regulation of mesenchymal cell proliferation involved in ureter development(GO:0072199) |
| 0.1 | 0.2 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.1 | 0.2 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.1 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.2 | GO:1904453 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.1 | 0.2 | GO:0002777 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) |
| 0.1 | 0.2 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 0.2 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.1 | 0.7 | GO:0070244 | negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.1 | 0.3 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.3 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.1 | 0.6 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 0.4 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.1 | 0.2 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.1 | 0.1 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 0.2 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.1 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.7 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.1 | 0.5 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.1 | 0.4 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 0.5 | GO:0002579 | positive regulation of antigen processing and presentation(GO:0002579) |
| 0.1 | 1.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.1 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.1 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 0.2 | GO:0097184 | amiloride transport(GO:0015898) cellular response to copper ion starvation(GO:0035874) response to azide(GO:0097184) cellular response to azide(GO:0097185) |
| 0.1 | 0.9 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 1.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 1.0 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.6 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.0 | 0.2 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.3 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.2 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.9 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.8 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.5 | GO:1903298 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.0 | 0.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.3 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.2 | GO:1900224 | zygotic specification of dorsal/ventral axis(GO:0007352) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.2 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 1.3 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.4 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.2 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.2 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.1 | GO:0061090 | positive regulation of sequestering of zinc ion(GO:0061090) |
| 0.0 | 0.5 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.6 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 1.3 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.2 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.1 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.0 | 0.1 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.0 | 0.1 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 | 0.2 | GO:0097393 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.0 | 0.4 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.2 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.2 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 1.0 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.2 | GO:0045964 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.5 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.2 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.2 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.8 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0014901 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.1 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.5 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.6 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.4 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.0 | 0.1 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.0 | 0.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.3 | GO:0060283 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.2 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.2 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 0.0 | 0.2 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.1 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.2 | GO:0042908 | exogenous drug catabolic process(GO:0042738) xenobiotic transport(GO:0042908) |
| 0.0 | 0.3 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.4 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.2 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.6 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.5 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.3 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.3 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 0.2 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.0 | 1.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.2 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.3 | GO:0090032 | negative regulation of steroid hormone biosynthetic process(GO:0090032) |
| 0.0 | 0.4 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.3 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.9 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.1 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.2 | GO:0060578 | subthalamic nucleus development(GO:0021763) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.3 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.2 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.5 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.1 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.0 | 0.2 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.4 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.4 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.3 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.2 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.2 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.0 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.0 | 1.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 1.2 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.0 | 0.1 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.0 | 1.9 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.0 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.3 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.5 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.4 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.2 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.0 | 0.2 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0070476 | rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.0 | 0.1 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.0 | 0.2 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0061037 | negative regulation of cartilage development(GO:0061037) |
| 0.0 | 0.1 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 1.3 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.3 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.0 | 0.0 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.5 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.0 | 0.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0045872 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.6 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.0 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.0 | 0.5 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:0090282 | positive regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0090282) |
| 0.0 | 0.1 | GO:1901738 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin A metabolic process(GO:1901738) |
| 0.0 | 0.2 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.1 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.1 | GO:0060690 | epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.0 | 0.1 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.3 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.1 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.6 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.7 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.1 | GO:1903208 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.0 | 0.3 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.3 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.1 | GO:0045348 | positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 0.0 | 1.2 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.2 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 | 0.4 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.0 | 0.1 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.6 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.1 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.2 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.2 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.0 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.0 | 0.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.1 | GO:0002461 | tolerance induction dependent upon immune response(GO:0002461) |
| 0.0 | 0.2 | GO:0046322 | negative regulation of fatty acid biosynthetic process(GO:0045717) negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.3 | GO:1903421 | regulation of synaptic vesicle recycling(GO:1903421) |
| 0.0 | 0.3 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.0 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.3 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.1 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.1 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.6 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.3 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.0 | 0.2 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.2 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.5 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.3 | GO:0002716 | negative regulation of natural killer cell mediated immunity(GO:0002716) |
| 0.0 | 0.2 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.1 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.0 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.0 | 0.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.7 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.3 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.1 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.3 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.2 | GO:0033574 | response to testosterone(GO:0033574) |
| 0.0 | 0.1 | GO:0043585 | nose morphogenesis(GO:0043585) alveolar primary septum development(GO:0061143) |
| 0.0 | 0.0 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.0 | 0.1 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.1 | GO:0002666 | positive regulation of T cell tolerance induction(GO:0002666) positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.6 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.3 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.6 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.4 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.0 | 0.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:1902510 | regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.0 | 1.9 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.6 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.1 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.3 | GO:0090218 | positive regulation of lipid kinase activity(GO:0090218) |
| 0.0 | 0.0 | GO:0009305 | protein biotinylation(GO:0009305) response to biotin(GO:0070781) histone biotinylation(GO:0071110) |
| 0.0 | 0.0 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.2 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.1 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.2 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.6 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.2 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.0 | 0.2 | GO:0030575 | nuclear body organization(GO:0030575) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 1.0 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.2 | 0.8 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.1 | 0.7 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 1.0 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 1.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 0.4 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 1.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 1.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 1.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 1.0 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.6 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.1 | 0.4 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 0.2 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 0.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.6 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.2 | GO:0016014 | dystrobrevin complex(GO:0016014) |
| 0.1 | 0.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.2 | GO:0097574 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.1 | 4.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.4 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.1 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.1 | 1.0 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 0.3 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.1 | 2.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 0.4 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 1.5 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 5.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.4 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.3 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 1.1 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.7 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 3.1 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.2 | GO:1990707 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.0 | 0.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.2 | GO:0032444 | activin responsive factor complex(GO:0032444) SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.5 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.5 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 2.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 1.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.3 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.1 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.4 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.0 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.2 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.2 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 2.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.6 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 1.0 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.3 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.3 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.3 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.8 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.4 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.2 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 1.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.0 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 3.5 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 2.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.5 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.5 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.2 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.7 | 2.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.6 | 4.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.6 | 4.8 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.5 | 1.6 | GO:0005009 | insulin-activated receptor activity(GO:0005009) |
| 0.5 | 1.5 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.5 | 2.0 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.5 | 1.4 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.4 | 7.5 | GO:0046977 | TAP binding(GO:0046977) |
| 0.4 | 2.0 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.4 | 2.2 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.4 | 1.8 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.3 | 3.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.3 | 0.9 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.3 | 1.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.3 | 1.9 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.3 | 3.0 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 0.7 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.2 | 1.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 1.4 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.2 | 2.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 1.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 1.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 0.7 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.2 | 0.9 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.2 | 0.5 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.2 | 1.0 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.2 | 0.5 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.2 | 1.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.2 | 5.3 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.2 | 0.6 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.2 | 0.5 | GO:0019002 | GMP binding(GO:0019002) |
| 0.2 | 1.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.2 | 0.9 | GO:0016807 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.6 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.6 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.4 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 0.4 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.1 | 0.7 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.1 | 1.0 | GO:0070404 | NADH binding(GO:0070404) |
| 0.1 | 0.5 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.1 | 1.9 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 1.5 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.5 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 0.8 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.4 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.1 | 0.3 | GO:0016855 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.1 | 1.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.6 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 0.7 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.1 | 0.3 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 0.6 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.3 | GO:0004771 | sterol esterase activity(GO:0004771) retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 0.4 | GO:0004844 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.1 | 0.5 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.4 | GO:0015205 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.1 | 0.9 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.6 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.6 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 0.5 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.3 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.5 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.2 | GO:0038052 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 0.4 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.4 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 0.4 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 1.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.2 | GO:0015152 | glucose-6-phosphate transmembrane transporter activity(GO:0015152) |
| 0.1 | 0.3 | GO:0019970 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.1 | 0.2 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.1 | 0.2 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.1 | 1.0 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.9 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 0.2 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 1.7 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 1.0 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.1 | 0.4 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.1 | 0.2 | GO:0047874 | dolichyldiphosphatase activity(GO:0047874) |
| 0.1 | 0.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.2 | GO:0008396 | oxysterol 7-alpha-hydroxylase activity(GO:0008396) |
| 0.1 | 0.3 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.4 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 0.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.2 | GO:0052600 | diamine oxidase activity(GO:0052597) histamine oxidase activity(GO:0052598) methylputrescine oxidase activity(GO:0052599) propane-1,3-diamine oxidase activity(GO:0052600) |
| 0.1 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.5 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.7 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.2 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 1.0 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.9 | GO:0070330 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) aromatase activity(GO:0070330) |
| 0.0 | 0.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 1.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.3 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 1.0 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.3 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.2 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.3 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.4 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.0 | 0.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.4 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.0 | GO:0071820 | N-box binding(GO:0071820) |
| 0.0 | 0.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.5 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0052796 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.7 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.7 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.2 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 1.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.6 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 1.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.2 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.0 | 0.3 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0004371 | glycerone kinase activity(GO:0004371) FAD-AMP lyase (cyclizing) activity(GO:0034012) triokinase activity(GO:0050354) |
| 0.0 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.3 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 1.1 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.5 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.7 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.6 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.1 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.0 | 1.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 1.5 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 1.5 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.5 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.3 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 1.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 1.0 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 2.0 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.2 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.1 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 2.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 1.0 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.2 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 2.2 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.0 | GO:0018271 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0016019 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.0 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 3.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 2.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 3.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 2.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 2.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.0 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.1 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.5 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.5 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 3.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 2.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 1.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 3.8 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.3 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.1 | 1.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 2.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 0.6 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.1 | 1.0 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 0.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.5 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 1.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 0.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 0.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.5 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 1.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 4.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.8 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.8 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.4 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 1.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.3 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.5 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.4 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.2 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 1.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |