Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Atf7_E4f1
Z-value: 0.83
Transcription factors associated with Atf7_E4f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
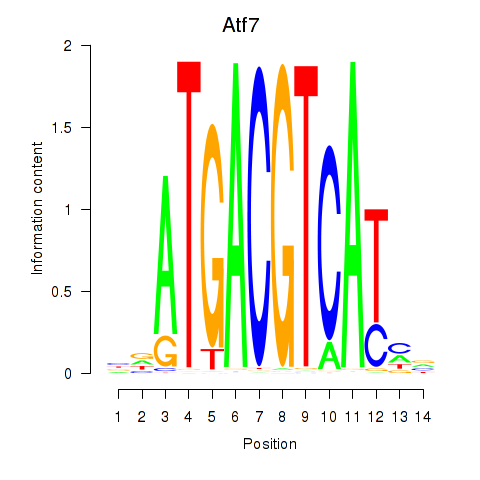
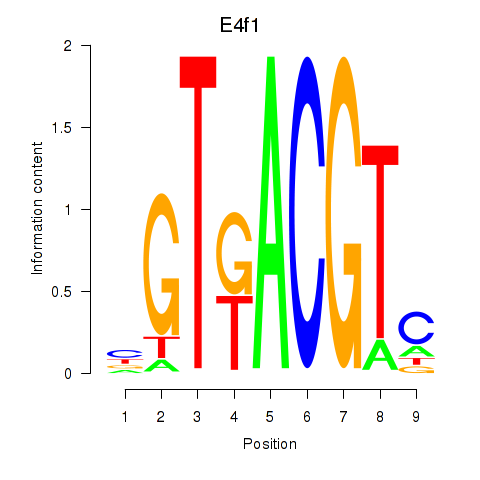
Atf7
|
ENSMUSG00000099083.8 | activating transcription factor 7 |
|
E4f1
|
ENSMUSG00000024137.10 | E4F transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E4f1 | mm39_v1_chr17_-_24674252_24674366 | 0.33 | 4.7e-02 | Click! |
| Atf7 | mm39_v1_chr15_-_102533494_102533535 | -0.22 | 2.1e-01 | Click! |
Activity profile of Atf7_E4f1 motif
Sorted Z-values of Atf7_E4f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_36626101 | 5.86 |
ENSMUST00000029270.10
|
Ccna2
|
cyclin A2 |
| chr5_+_123214332 | 4.27 |
ENSMUST00000067505.15
ENSMUST00000111619.10 ENSMUST00000160344.2 |
Tmem120b
|
transmembrane protein 120B |
| chr8_+_117648474 | 3.60 |
ENSMUST00000034205.5
ENSMUST00000212775.2 |
Cenpn
|
centromere protein N |
| chr5_+_143534455 | 3.26 |
ENSMUST00000169329.8
ENSMUST00000067145.12 ENSMUST00000119488.2 ENSMUST00000118121.2 ENSMUST00000200267.2 ENSMUST00000196487.2 |
Fam220a
Fam220a
|
family with sequence similarity 220, member A family with sequence similarity 220, member A |
| chr14_-_69522431 | 3.01 |
ENSMUST00000183882.2
ENSMUST00000037064.5 |
Slc25a37
|
solute carrier family 25, member 37 |
| chr11_+_113510135 | 2.90 |
ENSMUST00000146390.3
|
Sstr2
|
somatostatin receptor 2 |
| chr8_-_86091946 | 2.79 |
ENSMUST00000034133.14
|
Mylk3
|
myosin light chain kinase 3 |
| chr8_-_86091970 | 2.77 |
ENSMUST00000121972.8
|
Mylk3
|
myosin light chain kinase 3 |
| chr10_+_75407356 | 2.72 |
ENSMUST00000143226.8
ENSMUST00000124259.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr7_+_3706992 | 2.72 |
ENSMUST00000006496.15
ENSMUST00000108623.8 ENSMUST00000139818.2 ENSMUST00000108625.8 |
Rps9
|
ribosomal protein S9 |
| chr10_-_81214293 | 2.71 |
ENSMUST00000140901.8
|
Fzr1
|
fizzy and cell division cycle 20 related 1 |
| chr7_+_44117444 | 2.63 |
ENSMUST00000206887.2
ENSMUST00000117324.8 ENSMUST00000120852.8 ENSMUST00000134398.3 ENSMUST00000118628.8 |
Josd2
|
Josephin domain containing 2 |
| chr15_+_79231720 | 2.55 |
ENSMUST00000096350.11
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr14_-_69740670 | 2.50 |
ENSMUST00000180059.3
ENSMUST00000179116.3 |
Gm16867
|
predicted gene, 16867 |
| chr15_+_79232137 | 2.43 |
ENSMUST00000163691.3
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr15_+_98972850 | 2.42 |
ENSMUST00000039665.8
|
Troap
|
trophinin associated protein |
| chr7_+_44117511 | 2.32 |
ENSMUST00000121922.3
ENSMUST00000208117.2 |
Josd2
|
Josephin domain containing 2 |
| chr3_+_103078971 | 2.29 |
ENSMUST00000005830.15
|
Bcas2
|
breast carcinoma amplified sequence 2 |
| chr12_-_69205882 | 2.26 |
ENSMUST00000037023.9
|
Rps29
|
ribosomal protein S29 |
| chr7_+_44117404 | 2.20 |
ENSMUST00000035844.11
|
Josd2
|
Josephin domain containing 2 |
| chr7_+_44117475 | 2.17 |
ENSMUST00000118493.8
|
Josd2
|
Josephin domain containing 2 |
| chr6_-_86646118 | 2.17 |
ENSMUST00000001184.10
|
Mxd1
|
MAX dimerization protein 1 |
| chr3_+_89366425 | 2.15 |
ENSMUST00000029564.12
|
Pmvk
|
phosphomevalonate kinase |
| chr4_-_133972890 | 2.13 |
ENSMUST00000030644.8
|
Zfp593
|
zinc finger protein 593 |
| chr1_+_134383247 | 2.12 |
ENSMUST00000112232.8
ENSMUST00000027725.11 ENSMUST00000116528.2 |
Klhl12
|
kelch-like 12 |
| chr11_+_22940519 | 2.12 |
ENSMUST00000173867.8
|
Cct4
|
chaperonin containing Tcp1, subunit 4 (delta) |
| chr11_+_113510207 | 2.06 |
ENSMUST00000106630.2
|
Sstr2
|
somatostatin receptor 2 |
| chr8_-_124675939 | 2.02 |
ENSMUST00000044795.8
|
Nup133
|
nucleoporin 133 |
| chr7_-_133304244 | 1.95 |
ENSMUST00000209636.2
ENSMUST00000153698.3 |
Uros
|
uroporphyrinogen III synthase |
| chr3_+_40754489 | 1.93 |
ENSMUST00000203295.3
|
Plk4
|
polo like kinase 4 |
| chr2_-_126975804 | 1.90 |
ENSMUST00000110387.4
|
Ncaph
|
non-SMC condensin I complex, subunit H |
| chr2_+_164587901 | 1.87 |
ENSMUST00000017443.14
ENSMUST00000109326.10 |
Dnttip1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr4_+_130253925 | 1.86 |
ENSMUST00000105994.4
|
Snrnp40
|
small nuclear ribonucleoprotein 40 (U5) |
| chr1_-_134006847 | 1.82 |
ENSMUST00000020692.7
|
Btg2
|
BTG anti-proliferation factor 2 |
| chr12_+_103354915 | 1.82 |
ENSMUST00000101094.9
ENSMUST00000021620.13 |
Otub2
|
OTU domain, ubiquitin aldehyde binding 2 |
| chr11_+_22940599 | 1.82 |
ENSMUST00000020562.5
|
Cct4
|
chaperonin containing Tcp1, subunit 4 (delta) |
| chr16_-_57427179 | 1.81 |
ENSMUST00000114371.5
ENSMUST00000232413.2 |
Cmss1
|
cms small ribosomal subunit 1 |
| chr18_-_62313019 | 1.78 |
ENSMUST00000053640.5
|
Adrb2
|
adrenergic receptor, beta 2 |
| chr2_-_132095146 | 1.77 |
ENSMUST00000028817.7
|
Pcna
|
proliferating cell nuclear antigen |
| chr3_-_127346882 | 1.75 |
ENSMUST00000197668.2
ENSMUST00000029588.10 |
Larp7
|
La ribonucleoprotein domain family, member 7 |
| chr12_+_112611322 | 1.75 |
ENSMUST00000109755.5
|
Siva1
|
SIVA1, apoptosis-inducing factor |
| chr2_+_164587948 | 1.74 |
ENSMUST00000109327.4
|
Dnttip1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr11_-_78056347 | 1.73 |
ENSMUST00000017530.4
|
Traf4
|
TNF receptor associated factor 4 |
| chr7_-_141649003 | 1.73 |
ENSMUST00000039926.10
|
Dusp8
|
dual specificity phosphatase 8 |
| chr7_+_97437709 | 1.67 |
ENSMUST00000206984.2
|
Pak1
|
p21 (RAC1) activated kinase 1 |
| chr12_+_112611069 | 1.66 |
ENSMUST00000021728.12
|
Siva1
|
SIVA1, apoptosis-inducing factor |
| chr3_+_134918298 | 1.66 |
ENSMUST00000062893.12
|
Cenpe
|
centromere protein E |
| chr17_-_71833752 | 1.61 |
ENSMUST00000232863.2
ENSMUST00000024851.10 |
Ndc80
|
NDC80 kinetochore complex component |
| chr7_+_141027763 | 1.61 |
ENSMUST00000106003.2
|
Rplp2
|
ribosomal protein, large P2 |
| chr6_-_49240944 | 1.60 |
ENSMUST00000204189.3
ENSMUST00000031841.9 |
Tra2a
|
transformer 2 alpha |
| chr10_-_17823736 | 1.57 |
ENSMUST00000037879.8
|
Heca
|
hdc homolog, cell cycle regulator |
| chr9_-_96360676 | 1.54 |
ENSMUST00000057500.6
|
Rnf7
|
ring finger protein 7 |
| chr7_+_102090892 | 1.46 |
ENSMUST00000033283.10
|
Rrm1
|
ribonucleotide reductase M1 |
| chr5_+_33978035 | 1.46 |
ENSMUST00000075812.11
ENSMUST00000114397.9 ENSMUST00000155880.8 |
Nsd2
|
nuclear receptor binding SET domain protein 2 |
| chr12_+_4132567 | 1.39 |
ENSMUST00000020986.15
ENSMUST00000049584.6 |
Dnajc27
|
DnaJ heat shock protein family (Hsp40) member C27 |
| chr7_+_141027557 | 1.36 |
ENSMUST00000106004.8
|
Rplp2
|
ribosomal protein, large P2 |
| chr14_-_87378641 | 1.36 |
ENSMUST00000168889.3
ENSMUST00000022599.14 |
Diaph3
|
diaphanous related formin 3 |
| chr13_+_75855695 | 1.31 |
ENSMUST00000222194.2
ENSMUST00000223535.2 ENSMUST00000222853.2 |
Ell2
|
elongation factor for RNA polymerase II 2 |
| chr13_+_22227359 | 1.29 |
ENSMUST00000110452.2
|
H2bc11
|
H2B clustered histone 11 |
| chr4_-_131802561 | 1.28 |
ENSMUST00000105970.8
ENSMUST00000105975.8 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr10_-_30076543 | 1.25 |
ENSMUST00000099985.6
|
Cenpw
|
centromere protein W |
| chr9_+_20799471 | 1.25 |
ENSMUST00000004203.6
|
Ppan
|
peter pan homolog |
| chrX_-_8011952 | 1.25 |
ENSMUST00000115615.9
ENSMUST00000115616.8 ENSMUST00000115621.9 |
Rbm3
|
RNA binding motif (RNP1, RRM) protein 3 |
| chr6_-_30304512 | 1.25 |
ENSMUST00000094543.3
ENSMUST00000102993.10 |
Ube2h
|
ubiquitin-conjugating enzyme E2H |
| chr3_+_89366632 | 1.23 |
ENSMUST00000107410.8
|
Pmvk
|
phosphomevalonate kinase |
| chr4_-_117013396 | 1.22 |
ENSMUST00000102696.5
|
Rps8
|
ribosomal protein S8 |
| chr8_-_85567256 | 1.21 |
ENSMUST00000003911.13
ENSMUST00000109761.9 ENSMUST00000128035.2 |
Rad23a
|
RAD23 homolog A, nucleotide excision repair protein |
| chr17_-_37269425 | 1.20 |
ENSMUST00000172518.8
|
Polr1h
|
RNA polymerase I subunit H |
| chr9_-_96360648 | 1.20 |
ENSMUST00000071301.5
|
Rnf7
|
ring finger protein 7 |
| chr5_-_110801213 | 1.19 |
ENSMUST00000042147.6
|
Noc4l
|
NOC4 like |
| chr11_-_97673203 | 1.19 |
ENSMUST00000128801.2
ENSMUST00000103146.5 |
Rpl23
|
ribosomal protein L23 |
| chr11_-_40624200 | 1.18 |
ENSMUST00000020579.9
|
Hmmr
|
hyaluronan mediated motility receptor (RHAMM) |
| chr7_+_45367479 | 1.16 |
ENSMUST00000072503.13
ENSMUST00000211061.2 ENSMUST00000209693.2 |
Rpl18
|
ribosomal protein L18 |
| chr10_-_111833138 | 1.15 |
ENSMUST00000074805.12
|
Glipr1
|
GLI pathogenesis-related 1 (glioma) |
| chr15_+_79975520 | 1.14 |
ENSMUST00000009728.13
ENSMUST00000009727.12 |
Syngr1
|
synaptogyrin 1 |
| chr7_+_45367545 | 1.14 |
ENSMUST00000209287.2
|
Rpl18
|
ribosomal protein L18 |
| chrX_-_8011918 | 1.11 |
ENSMUST00000115619.8
ENSMUST00000115617.10 ENSMUST00000040010.10 |
Rbm3
|
RNA binding motif (RNP1, RRM) protein 3 |
| chr17_-_37269330 | 1.09 |
ENSMUST00000113669.9
|
Polr1h
|
RNA polymerase I subunit H |
| chr6_-_25809209 | 1.09 |
ENSMUST00000115330.8
|
Pot1a
|
protection of telomeres 1A |
| chr17_+_32255071 | 1.09 |
ENSMUST00000081339.13
|
Rrp1b
|
ribosomal RNA processing 1B |
| chr7_-_30364394 | 1.08 |
ENSMUST00000019697.9
|
Haus5
|
HAUS augmin-like complex, subunit 5 |
| chrX_+_134894573 | 1.06 |
ENSMUST00000058119.9
|
Arxes2
|
adipocyte-related X-chromosome expressed sequence 2 |
| chr15_-_38519499 | 1.05 |
ENSMUST00000110329.8
ENSMUST00000065308.13 |
Azin1
|
antizyme inhibitor 1 |
| chr8_+_67312847 | 1.03 |
ENSMUST00000118009.2
|
Naf1
|
nuclear assembly factor 1 ribonucleoprotein |
| chr9_+_54606144 | 1.03 |
ENSMUST00000120452.8
|
Dnaja4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr1_-_75455915 | 1.02 |
ENSMUST00000079205.14
ENSMUST00000094818.4 |
Chpf
|
chondroitin polymerizing factor |
| chr8_-_26087475 | 1.02 |
ENSMUST00000210810.2
ENSMUST00000210616.2 ENSMUST00000079160.8 |
Letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr4_-_131802606 | 1.01 |
ENSMUST00000146021.8
|
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr5_+_33176160 | 0.98 |
ENSMUST00000019109.8
|
Ywhah
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide |
| chr8_+_123829089 | 0.98 |
ENSMUST00000000756.6
|
Rpl13
|
ribosomal protein L13 |
| chr17_-_34043502 | 0.97 |
ENSMUST00000087342.13
ENSMUST00000173844.8 |
Rps28
|
ribosomal protein S28 |
| chr16_+_49519561 | 0.96 |
ENSMUST00000046777.11
ENSMUST00000142682.9 |
Ift57
|
intraflagellar transport 57 |
| chr7_+_30113170 | 0.95 |
ENSMUST00000207982.2
ENSMUST00000032800.10 |
Tyrobp
|
TYRO protein tyrosine kinase binding protein |
| chr13_-_21937997 | 0.95 |
ENSMUST00000074752.4
|
H2ac15
|
H2A clustered histone 15 |
| chr6_-_131270136 | 0.94 |
ENSMUST00000032307.12
|
Magohb
|
mago homolog B, exon junction complex core component |
| chr2_+_109111083 | 0.94 |
ENSMUST00000028527.8
|
Kif18a
|
kinesin family member 18A |
| chr12_+_73333641 | 0.92 |
ENSMUST00000153941.8
ENSMUST00000122920.8 ENSMUST00000101313.4 |
Slc38a6
|
solute carrier family 38, member 6 |
| chr17_+_26882171 | 0.91 |
ENSMUST00000236346.2
|
Atp6v0e
|
ATPase, H+ transporting, lysosomal V0 subunit E |
| chr17_-_34043320 | 0.91 |
ENSMUST00000173879.8
ENSMUST00000166693.3 ENSMUST00000173019.8 |
Rps28
|
ribosomal protein S28 |
| chr11_+_69805005 | 0.90 |
ENSMUST00000057884.6
|
Gps2
|
G protein pathway suppressor 2 |
| chr17_-_26727437 | 0.87 |
ENSMUST00000236661.2
ENSMUST00000025025.7 |
Dusp1
|
dual specificity phosphatase 1 |
| chr3_+_40754448 | 0.87 |
ENSMUST00000026858.11
|
Plk4
|
polo like kinase 4 |
| chr2_-_156848923 | 0.86 |
ENSMUST00000146413.8
ENSMUST00000103129.9 ENSMUST00000103130.8 |
Dsn1
|
DSN1 homolog, MIS12 kinetochore complex component |
| chr7_-_105393519 | 0.86 |
ENSMUST00000141116.2
|
Taf10
|
TATA-box binding protein associated factor 10 |
| chr14_-_67309866 | 0.86 |
ENSMUST00000225380.2
ENSMUST00000089230.7 |
Ppp2r2a
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr13_+_8935537 | 0.84 |
ENSMUST00000169314.9
|
Idi1
|
isopentenyl-diphosphate delta isomerase |
| chr7_-_44635813 | 0.84 |
ENSMUST00000208829.2
ENSMUST00000207370.2 ENSMUST00000107843.11 |
Prmt1
|
protein arginine N-methyltransferase 1 |
| chr6_+_113508636 | 0.83 |
ENSMUST00000036340.12
ENSMUST00000204827.3 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr19_-_5962798 | 0.82 |
ENSMUST00000118623.2
|
Dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr1_+_59724108 | 0.82 |
ENSMUST00000027174.10
ENSMUST00000190231.7 ENSMUST00000191142.7 ENSMUST00000185772.7 |
Nop58
|
NOP58 ribonucleoprotein |
| chr13_-_9046020 | 0.82 |
ENSMUST00000222098.2
|
Gtpbp4
|
GTP binding protein 4 |
| chr19_-_24938909 | 0.81 |
ENSMUST00000025815.10
|
Cbwd1
|
COBW domain containing 1 |
| chr12_+_108572015 | 0.81 |
ENSMUST00000109854.9
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chr10_-_128361731 | 0.81 |
ENSMUST00000026427.8
|
Esyt1
|
extended synaptotagmin-like protein 1 |
| chr7_-_44635740 | 0.81 |
ENSMUST00000209056.3
ENSMUST00000209124.2 ENSMUST00000208312.2 ENSMUST00000207659.2 ENSMUST00000045325.14 |
Prmt1
|
protein arginine N-methyltransferase 1 |
| chr14_+_47710005 | 0.80 |
ENSMUST00000043112.9
ENSMUST00000163324.8 ENSMUST00000228668.2 ENSMUST00000168833.9 |
Fbxo34
|
F-box protein 34 |
| chr8_+_75836187 | 0.79 |
ENSMUST00000164309.3
ENSMUST00000212426.2 ENSMUST00000212811.2 |
Mcm5
|
minichromosome maintenance complex component 5 |
| chr5_+_101912939 | 0.76 |
ENSMUST00000031273.9
|
Cds1
|
CDP-diacylglycerol synthase 1 |
| chr11_-_86999481 | 0.75 |
ENSMUST00000051395.9
|
Prr11
|
proline rich 11 |
| chr14_+_47710085 | 0.75 |
ENSMUST00000095941.9
|
Fbxo34
|
F-box protein 34 |
| chr18_+_82932747 | 0.74 |
ENSMUST00000071233.7
|
Zfp516
|
zinc finger protein 516 |
| chr13_-_100881117 | 0.74 |
ENSMUST00000078573.5
ENSMUST00000109333.8 |
Mrps36
|
mitochondrial ribosomal protein S36 |
| chr12_+_102710008 | 0.74 |
ENSMUST00000057416.8
|
Tmem251
|
transmembrane protein 251 |
| chr11_+_68792364 | 0.73 |
ENSMUST00000073471.13
ENSMUST00000101014.9 ENSMUST00000128952.8 ENSMUST00000167436.3 |
Rpl26
|
ribosomal protein L26 |
| chr5_+_114582327 | 0.73 |
ENSMUST00000137167.8
ENSMUST00000112239.9 ENSMUST00000124260.8 ENSMUST00000125650.6 ENSMUST00000043760.15 |
Mvk
|
mevalonate kinase |
| chr8_-_4375320 | 0.73 |
ENSMUST00000098950.6
|
Elavl1
|
ELAV (embryonic lethal, abnormal vision)-like 1 (Hu antigen R) |
| chr1_+_175708341 | 0.71 |
ENSMUST00000195196.6
ENSMUST00000194306.6 ENSMUST00000193822.6 |
Exo1
|
exonuclease 1 |
| chr7_+_127503251 | 0.71 |
ENSMUST00000071056.14
|
Bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr16_-_31984272 | 0.70 |
ENSMUST00000231836.2
ENSMUST00000115163.4 ENSMUST00000144345.2 ENSMUST00000143682.8 ENSMUST00000115165.10 ENSMUST00000099991.11 ENSMUST00000130410.8 |
Nrros
|
negative regulator of reactive oxygen species |
| chr13_+_38220971 | 0.70 |
ENSMUST00000021866.10
|
Riok1
|
RIO kinase 1 |
| chr8_+_71151581 | 0.69 |
ENSMUST00000095267.8
|
Jund
|
jun D proto-oncogene |
| chr16_+_21828223 | 0.69 |
ENSMUST00000023561.8
|
Senp2
|
SUMO/sentrin specific peptidase 2 |
| chr8_-_23143422 | 0.69 |
ENSMUST00000033938.7
|
Polb
|
polymerase (DNA directed), beta |
| chr5_+_24369961 | 0.69 |
ENSMUST00000049887.13
|
Nupl2
|
nucleoporin like 2 |
| chr10_-_128383508 | 0.68 |
ENSMUST00000152539.8
ENSMUST00000133458.2 ENSMUST00000040572.10 |
Zc3h10
|
zinc finger CCCH type containing 10 |
| chr8_-_81466126 | 0.68 |
ENSMUST00000043359.9
|
Smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr2_-_163486998 | 0.68 |
ENSMUST00000017851.4
|
Serinc3
|
serine incorporator 3 |
| chr14_-_30741012 | 0.68 |
ENSMUST00000037739.8
|
Gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr17_-_37269153 | 0.67 |
ENSMUST00000172823.2
|
Polr1h
|
RNA polymerase I subunit H |
| chr11_-_88608920 | 0.67 |
ENSMUST00000092794.12
|
Msi2
|
musashi RNA-binding protein 2 |
| chr9_-_59393893 | 0.66 |
ENSMUST00000171975.8
|
Arih1
|
ariadne RBR E3 ubiquitin protein ligase 1 |
| chr15_+_43340609 | 0.66 |
ENSMUST00000022962.8
|
Emc2
|
ER membrane protein complex subunit 2 |
| chr10_+_72490705 | 0.65 |
ENSMUST00000105431.8
ENSMUST00000160337.2 |
Zwint
|
ZW10 interactor |
| chr13_+_120151982 | 0.65 |
ENSMUST00000179869.3
ENSMUST00000224188.2 |
Hmgcs1
|
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 |
| chr11_+_87000032 | 0.65 |
ENSMUST00000020794.6
|
Ska2
|
spindle and kinetochore associated complex subunit 2 |
| chr17_-_35055843 | 0.64 |
ENSMUST00000077477.12
|
Stk19
|
serine/threonine kinase 19 |
| chr10_-_78248771 | 0.64 |
ENSMUST00000062678.11
|
Rrp1
|
ribosomal RNA processing 1 |
| chr5_-_130284366 | 0.63 |
ENSMUST00000026387.11
|
Sbds
|
SBDS ribosome maturation factor |
| chr12_+_79177523 | 0.62 |
ENSMUST00000021550.7
|
Arg2
|
arginase type II |
| chr18_+_11790409 | 0.62 |
ENSMUST00000047322.8
|
Rbbp8
|
retinoblastoma binding protein 8, endonuclease |
| chr11_-_70128678 | 0.61 |
ENSMUST00000108575.9
|
0610010K14Rik
|
RIKEN cDNA 0610010K14 gene |
| chr10_+_72490677 | 0.61 |
ENSMUST00000020081.11
|
Zwint
|
ZW10 interactor |
| chr6_+_54658609 | 0.61 |
ENSMUST00000190641.7
ENSMUST00000187701.2 |
Mturn
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr2_-_132420074 | 0.60 |
ENSMUST00000110136.8
ENSMUST00000124107.8 ENSMUST00000060955.12 |
Gpcpd1
|
glycerophosphocholine phosphodiesterase 1 |
| chr11_+_59099147 | 0.60 |
ENSMUST00000020719.7
|
2310033P09Rik
|
RIKEN cDNA 2310033P09 gene |
| chr7_-_103778992 | 0.59 |
ENSMUST00000053743.6
|
Ubqln5
|
ubiquilin 5 |
| chr2_-_124965537 | 0.59 |
ENSMUST00000142718.8
ENSMUST00000152367.8 ENSMUST00000067780.10 ENSMUST00000147105.8 |
Myef2
|
myelin basic protein expression factor 2, repressor |
| chr13_-_96807346 | 0.59 |
ENSMUST00000022176.15
|
Hmgcr
|
3-hydroxy-3-methylglutaryl-Coenzyme A reductase |
| chr6_-_25809188 | 0.59 |
ENSMUST00000115327.8
|
Pot1a
|
protection of telomeres 1A |
| chr19_-_4675352 | 0.58 |
ENSMUST00000224707.2
ENSMUST00000237059.2 |
Rce1
|
Ras converting CAAX endopeptidase 1 |
| chr5_-_38719025 | 0.58 |
ENSMUST00000005234.13
|
Wdr1
|
WD repeat domain 1 |
| chr11_-_101442663 | 0.58 |
ENSMUST00000017290.11
|
Brca1
|
breast cancer 1, early onset |
| chr11_+_100211363 | 0.58 |
ENSMUST00000152521.2
|
Eif1
|
eukaryotic translation initiation factor 1 |
| chr13_+_21938258 | 0.58 |
ENSMUST00000091709.3
|
H2bc15
|
H2B clustered histone 15 |
| chr11_-_69563133 | 0.57 |
ENSMUST00000163666.3
|
Eif4a1
|
eukaryotic translation initiation factor 4A1 |
| chr4_-_46138397 | 0.57 |
ENSMUST00000144495.2
ENSMUST00000107770.2 ENSMUST00000156021.2 ENSMUST00000107772.8 |
Tstd2
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 2 |
| chr19_-_61215743 | 0.56 |
ENSMUST00000237386.2
|
Csf2ra
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr11_-_59054521 | 0.56 |
ENSMUST00000137433.2
ENSMUST00000054523.6 |
Iba57
|
IBA57 homolog, iron-sulfur cluster assembly |
| chr15_-_79571977 | 0.55 |
ENSMUST00000023061.7
|
Josd1
|
Josephin domain containing 1 |
| chr9_+_64718708 | 0.55 |
ENSMUST00000213926.2
|
Dennd4a
|
DENN/MADD domain containing 4A |
| chr11_+_76134541 | 0.55 |
ENSMUST00000040577.5
|
Mrm3
|
mitochondrial rRNA methyltransferase 3 |
| chr1_+_75145275 | 0.55 |
ENSMUST00000162768.8
ENSMUST00000160439.8 ENSMUST00000027394.12 |
Zfand2b
|
zinc finger, AN1 type domain 2B |
| chr5_+_137628377 | 0.55 |
ENSMUST00000175968.8
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr16_+_21613068 | 0.54 |
ENSMUST00000211443.2
ENSMUST00000231300.2 ENSMUST00000209449.2 ENSMUST00000181780.9 ENSMUST00000209728.2 ENSMUST00000181960.3 ENSMUST00000209429.2 ENSMUST00000180830.3 ENSMUST00000231988.2 |
1300002E11Rik
Map3k13
|
RIKEN cDNA 1300002E11 gene mitogen-activated protein kinase kinase kinase 13 |
| chr7_+_44397837 | 0.54 |
ENSMUST00000002275.15
|
Vrk3
|
vaccinia related kinase 3 |
| chr17_-_28126679 | 0.53 |
ENSMUST00000025057.5
|
Taf11
|
TATA-box binding protein associated factor 11 |
| chr11_-_70128712 | 0.53 |
ENSMUST00000108577.8
ENSMUST00000108579.8 ENSMUST00000021181.13 ENSMUST00000108578.9 ENSMUST00000102569.10 |
0610010K14Rik
|
RIKEN cDNA 0610010K14 gene |
| chr2_+_120439858 | 0.53 |
ENSMUST00000124187.8
|
Haus2
|
HAUS augmin-like complex, subunit 2 |
| chr9_+_20914012 | 0.52 |
ENSMUST00000003386.7
|
Mrpl4
|
mitochondrial ribosomal protein L4 |
| chr7_+_30411634 | 0.52 |
ENSMUST00000005692.14
ENSMUST00000170371.2 |
Atp4a
|
ATPase, H+/K+ exchanging, gastric, alpha polypeptide |
| chr1_-_165064455 | 0.52 |
ENSMUST00000043235.8
|
Tiprl
|
TIP41, TOR signalling pathway regulator-like (S. cerevisiae) |
| chr19_+_36903471 | 0.51 |
ENSMUST00000099494.4
|
Btaf1
|
B-TFIID TATA-box binding protein associated factor 1 |
| chr17_-_57318271 | 0.51 |
ENSMUST00000002733.7
|
Gtf2f1
|
general transcription factor IIF, polypeptide 1 |
| chr11_+_110858842 | 0.51 |
ENSMUST00000180023.8
ENSMUST00000106636.8 |
Kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr14_+_51045298 | 0.51 |
ENSMUST00000036126.7
|
Parp2
|
poly (ADP-ribose) polymerase family, member 2 |
| chr11_+_68582731 | 0.51 |
ENSMUST00000102611.10
|
Myh10
|
myosin, heavy polypeptide 10, non-muscle |
| chr4_+_46138576 | 0.51 |
ENSMUST00000030014.9
|
Ncbp1
|
nuclear cap binding protein subunit 1 |
| chr1_-_191307648 | 0.50 |
ENSMUST00000027933.11
|
Dtl
|
denticleless E3 ubiquitin protein ligase |
| chr19_-_61216834 | 0.49 |
ENSMUST00000076046.7
|
Csf2ra
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr4_-_151142351 | 0.49 |
ENSMUST00000030797.4
|
Vamp3
|
vesicle-associated membrane protein 3 |
| chr2_-_36026614 | 0.49 |
ENSMUST00000122456.8
|
Rbm18
|
RNA binding motif protein 18 |
| chr17_+_35201130 | 0.49 |
ENSMUST00000173004.2
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr9_+_20914211 | 0.48 |
ENSMUST00000214124.2
ENSMUST00000216818.2 |
Mrpl4
|
mitochondrial ribosomal protein L4 |
| chr5_+_150446064 | 0.48 |
ENSMUST00000044620.11
ENSMUST00000202003.4 |
Brca2
|
breast cancer 2, early onset |
| chr10_-_128329693 | 0.48 |
ENSMUST00000217776.2
ENSMUST00000219236.2 ENSMUST00000217733.2 ENSMUST00000218127.2 ENSMUST00000217969.2 ENSMUST00000164181.2 |
Myl6
|
myosin, light polypeptide 6, alkali, smooth muscle and non-muscle |
| chr15_-_51728893 | 0.48 |
ENSMUST00000022925.10
|
Eif3h
|
eukaryotic translation initiation factor 3, subunit H |
| chr7_+_90091937 | 0.47 |
ENSMUST00000061767.5
|
Crebzf
|
CREB/ATF bZIP transcription factor |
| chr7_-_101859379 | 0.47 |
ENSMUST00000210682.2
|
Nup98
|
nucleoporin 98 |
| chr10_+_79518002 | 0.47 |
ENSMUST00000020550.13
|
Cdc34
|
cell division cycle 34 |
| chr3_+_127347099 | 0.46 |
ENSMUST00000043108.9
ENSMUST00000196141.5 ENSMUST00000195955.2 |
Zgrf1
|
zinc finger, GRF-type containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Atf7_E4f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.9 | 5.6 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.6 | 1.9 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.5 | 3.9 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.5 | 1.9 | GO:2000371 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.5 | 1.4 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.4 | 1.8 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.4 | 1.8 | GO:1902990 | leading strand elongation(GO:0006272) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.4 | 1.7 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.4 | 1.7 | GO:0051316 | attachment of spindle microtubules to kinetochore involved in meiotic chromosome segregation(GO:0051316) |
| 0.4 | 1.5 | GO:0003290 | atrial septum secundum morphogenesis(GO:0003290) |
| 0.4 | 1.4 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.4 | 2.8 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.3 | 2.7 | GO:1901748 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.3 | 2.7 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.3 | 1.6 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.3 | 1.0 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.3 | 1.3 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.3 | 5.0 | GO:0030432 | peristalsis(GO:0030432) |
| 0.3 | 1.2 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.3 | 1.2 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.3 | 2.0 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.3 | 1.0 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.3 | 0.8 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) negative regulation of histone phosphorylation(GO:0033128) |
| 0.3 | 3.6 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.2 | 1.2 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.2 | 2.9 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.2 | 2.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.2 | 3.3 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.2 | 0.7 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.2 | 1.0 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.2 | 3.3 | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901030) |
| 0.2 | 2.3 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.2 | 0.7 | GO:1902164 | positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.2 | 0.5 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.2 | 0.7 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.2 | 0.5 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.2 | 0.6 | GO:0010958 | regulation of amino acid import(GO:0010958) |
| 0.2 | 0.8 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.2 | 5.1 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.1 | 0.4 | GO:2000847 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.1 | 1.3 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 2.7 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 0.7 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 0.1 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.1 | 1.9 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.5 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 2.3 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 1.3 | GO:0007549 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 | 0.8 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 1.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.4 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.1 | 0.8 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 0.9 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 1.0 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.4 | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 0.1 | 5.7 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 0.6 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 1.4 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 2.5 | GO:0031442 | positive regulation of mRNA 3'-end processing(GO:0031442) |
| 0.1 | 1.1 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.1 | 0.5 | GO:0021763 | subthalamic nucleus development(GO:0021763) superior vena cava morphogenesis(GO:0060578) |
| 0.1 | 0.3 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.1 | 0.2 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) |
| 0.1 | 0.7 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 0.6 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 1.7 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 1.7 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.5 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.2 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.2 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 0.2 | GO:0051385 | response to mineralocorticoid(GO:0051385) |
| 0.1 | 0.2 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.1 | 0.2 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 | 4.5 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.1 | 0.8 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.9 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.1 | 0.2 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.1 | 1.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 1.5 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 0.3 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.9 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.1 | 0.2 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 0.4 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 3.8 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.3 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.1 | 3.1 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 3.0 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 1.1 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.1 | 0.2 | GO:1990173 | protein localization to nucleoplasm(GO:1990173) |
| 0.1 | 0.2 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 | 0.8 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.2 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 0.2 | GO:0002191 | formation of cytoplasmic translation initiation complex(GO:0001732) cap-dependent translational initiation(GO:0002191) |
| 0.1 | 0.5 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.6 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.1 | 0.7 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.5 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.7 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 1.3 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.7 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 | 0.1 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.8 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 6.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 1.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.9 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 | 0.1 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.0 | 0.2 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.0 | 0.2 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 1.5 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.2 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.3 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 1.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 1.0 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.8 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 1.2 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.2 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.2 | GO:0060702 | regulation of endoribonuclease activity(GO:0060699) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) |
| 0.0 | 0.5 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.4 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.5 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.7 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.4 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.2 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 3.9 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.0 | 0.1 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.0 | 0.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 1.2 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.5 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.9 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.9 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 | 0.4 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.7 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.2 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.4 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.5 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.5 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.3 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.5 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.2 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.5 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.9 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.3 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.0 | 0.5 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.6 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 1.1 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 0.9 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 2.6 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.3 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.6 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.3 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.0 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.0 | 0.0 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.2 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.2 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.3 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.3 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 | 1.3 | GO:0035308 | negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.4 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 3.6 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 1.5 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.2 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) motor behavior(GO:0061744) |
| 0.0 | 4.6 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 0.0 | GO:0061187 | histone H4-K20 demethylation(GO:0035574) regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.1 | GO:0046878 | angiotensin-mediated drinking behavior(GO:0003051) positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.4 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.5 | 2.7 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.5 | 5.9 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.4 | 1.6 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.4 | 2.8 | GO:0098536 | deuterosome(GO:0098536) |
| 0.4 | 4.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.4 | 1.9 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.3 | 0.9 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.3 | 3.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.2 | 1.5 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.2 | 1.4 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.2 | 3.6 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.2 | 0.6 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.2 | 1.8 | GO:0070187 | telosome(GO:0070187) |
| 0.2 | 1.6 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 0.9 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.2 | 2.9 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.2 | 2.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.7 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 8.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.6 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.9 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.3 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 2.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 8.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 2.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 3.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.8 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 1.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.5 | GO:0044393 | microspike(GO:0044393) |
| 0.1 | 0.8 | GO:0070761 | box C/D snoRNP complex(GO:0031428) pre-snoRNP complex(GO:0070761) |
| 0.1 | 1.0 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 4.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 0.7 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 1.9 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 2.0 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 1.7 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 0.5 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 1.0 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.1 | 1.0 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.7 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 1.0 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.1 | 0.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 6.8 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 1.0 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.6 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 3.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 2.0 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.5 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.1 | GO:0002095 | caveolar macromolecular signaling complex(GO:0002095) |
| 0.0 | 1.1 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.9 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.7 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.2 | GO:0030532 | small nuclear ribonucleoprotein complex(GO:0030532) |
| 0.0 | 1.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.8 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 1.2 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.2 | GO:0045240 | dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.5 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 3.8 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.2 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.5 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.8 | 5.0 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.7 | 2.7 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.6 | 5.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.6 | 1.8 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) dinucleotide insertion or deletion binding(GO:0032139) |
| 0.6 | 1.7 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.5 | 4.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.5 | 2.7 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.4 | 1.8 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.4 | 1.6 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.4 | 1.4 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.4 | 1.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.3 | 1.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.3 | 1.9 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.3 | 0.9 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.3 | 1.9 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.3 | 1.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.3 | 1.8 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.3 | 0.8 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.2 | 2.7 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.2 | 1.0 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.2 | 1.2 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.2 | 1.5 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.2 | 0.7 | GO:0004420 | hydroxymethylglutaryl-CoA reductase (NADPH) activity(GO:0004420) hydroxymethylglutaryl-CoA reductase activity(GO:0042282) |
| 0.2 | 1.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 1.7 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 2.7 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.2 | 0.6 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.2 | 0.5 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 0.2 | 2.7 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.2 | 0.5 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.2 | 0.8 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 0.7 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.1 | 0.4 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 3.5 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 1.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 6.2 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 1.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.3 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 3.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 2.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.3 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.1 | 0.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.2 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 0.8 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 0.7 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.6 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.1 | 0.6 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 2.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 7.2 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 12.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 1.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.7 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 1.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 4.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 0.7 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.6 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 1.0 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 1.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 1.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 2.1 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 3.4 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.1 | 0.7 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.5 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 0.5 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.7 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 2.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.4 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.3 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 1.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.2 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.6 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.2 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 1.0 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.2 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.9 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 1.0 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 1.0 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0008821 | recombinase activity(GO:0000150) crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 1.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.3 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 1.5 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 4.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.7 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.4 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 1.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.9 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 1.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.0 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.0 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 2.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 3.9 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 6.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 2.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 2.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.5 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 1.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.3 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.8 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.5 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 1.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.1 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.9 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.7 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.5 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.4 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.9 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 6.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 4.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.8 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 8.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.0 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 8.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 2.9 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 1.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 2.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 3.1 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 1.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 3.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 0.7 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 2.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 3.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 4.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.0 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.8 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.4 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.9 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.6 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 0.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.9 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 3.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.1 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.7 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 3.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.7 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.2 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |