Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Atoh1_Bhlhe23
Z-value: 0.41
Transcription factors associated with Atoh1_Bhlhe23
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
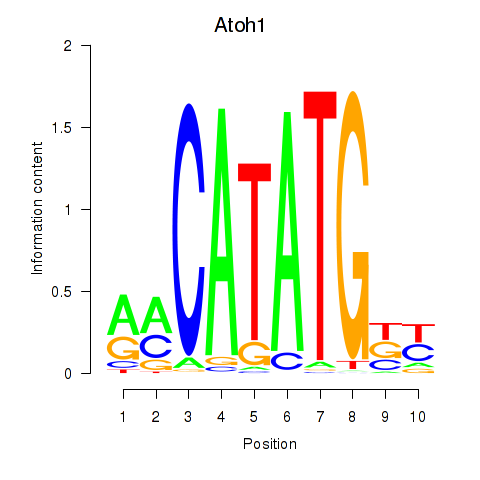
Atoh1
|
ENSMUSG00000073043.6 | atonal bHLH transcription factor 1 |
|
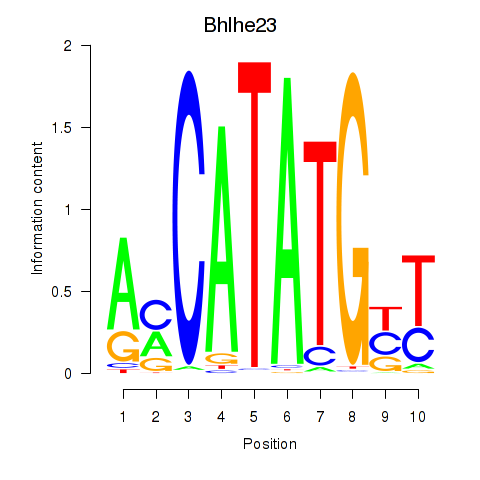
Bhlhe23
|
ENSMUSG00000045493.5 | basic helix-loop-helix family, member e23 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atoh1 | mm39_v1_chr6_+_64706101_64706117 | 0.19 | 2.7e-01 | Click! |
Activity profile of Atoh1_Bhlhe23 motif
Sorted Z-values of Atoh1_Bhlhe23 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_70945434 | 3.43 |
ENSMUST00000228346.2
|
Xpo7
|
exportin 7 |
| chr7_+_127503812 | 3.26 |
ENSMUST00000151451.3
ENSMUST00000124533.3 ENSMUST00000206745.2 ENSMUST00000206140.2 |
Bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr14_+_43951187 | 2.55 |
ENSMUST00000094051.6
|
Gm7324
|
predicted gene 7324 |
| chr7_-_126303947 | 2.37 |
ENSMUST00000032949.14
|
Coro1a
|
coronin, actin binding protein 1A |
| chrX_+_55500170 | 1.96 |
ENSMUST00000039374.9
ENSMUST00000101553.9 ENSMUST00000186445.7 |
Ints6l
|
integrator complex subunit 6 like |
| chr8_-_107792264 | 1.94 |
ENSMUST00000034393.7
|
Tmed6
|
transmembrane p24 trafficking protein 6 |
| chr7_-_126303887 | 1.81 |
ENSMUST00000131415.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr10_-_128237087 | 1.77 |
ENSMUST00000042666.13
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr10_-_128236317 | 1.76 |
ENSMUST00000167859.2
ENSMUST00000218858.2 |
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr7_-_126736979 | 1.76 |
ENSMUST00000049931.6
|
Spn
|
sialophorin |
| chr9_-_21829385 | 1.73 |
ENSMUST00000128442.2
ENSMUST00000119055.8 ENSMUST00000122211.8 ENSMUST00000115351.10 |
Rab3d
|
RAB3D, member RAS oncogene family |
| chr11_-_69493567 | 1.67 |
ENSMUST00000138694.2
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide |
| chr10_-_128236366 | 1.58 |
ENSMUST00000219131.2
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr9_+_56344700 | 1.57 |
ENSMUST00000239472.2
|
ENSMUSG00000118653.2
|
ubiquitin-conjugating enzyme E2S (Ube2s) retrogene |
| chr11_-_97673203 | 1.26 |
ENSMUST00000128801.2
ENSMUST00000103146.5 |
Rpl23
|
ribosomal protein L23 |
| chrX_+_70599524 | 1.26 |
ENSMUST00000072699.13
ENSMUST00000114582.9 ENSMUST00000015361.11 ENSMUST00000088874.10 |
Hmgb3
|
high mobility group box 3 |
| chr7_-_126303689 | 1.25 |
ENSMUST00000135087.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr19_-_46033353 | 1.15 |
ENSMUST00000026252.14
ENSMUST00000156585.9 ENSMUST00000185355.7 ENSMUST00000152946.8 |
Ldb1
|
LIM domain binding 1 |
| chr3_-_126792056 | 1.15 |
ENSMUST00000044443.15
|
Ank2
|
ankyrin 2, brain |
| chr19_-_53577499 | 1.13 |
ENSMUST00000095978.5
|
Nutf2-ps1
|
nuclear transport factor 2, pseudogene 1 |
| chr9_-_71803354 | 0.98 |
ENSMUST00000184448.8
|
Tcf12
|
transcription factor 12 |
| chr3_+_87814147 | 0.94 |
ENSMUST00000159492.8
|
Hdgf
|
heparin binding growth factor |
| chr11_+_58845502 | 0.93 |
ENSMUST00000108817.5
ENSMUST00000047697.12 |
H2aw
Trim17
|
H2A.W histone tripartite motif-containing 17 |
| chr11_-_72441054 | 0.89 |
ENSMUST00000021154.7
|
Spns3
|
spinster homolog 3 |
| chr7_+_127503251 | 0.86 |
ENSMUST00000071056.14
|
Bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr10_-_25076008 | 0.85 |
ENSMUST00000100012.3
|
Akap7
|
A kinase (PRKA) anchor protein 7 |
| chr11_+_31822211 | 0.78 |
ENSMUST00000020543.13
ENSMUST00000109412.9 |
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr3_+_3573084 | 0.77 |
ENSMUST00000108393.8
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr7_+_28488380 | 0.77 |
ENSMUST00000209035.2
ENSMUST00000059857.8 |
Rinl
|
Ras and Rab interactor-like |
| chr6_-_119925387 | 0.76 |
ENSMUST00000162541.8
|
Wnk1
|
WNK lysine deficient protein kinase 1 |
| chr16_+_57173456 | 0.75 |
ENSMUST00000159816.8
|
Filip1l
|
filamin A interacting protein 1-like |
| chr5_+_3393893 | 0.73 |
ENSMUST00000165117.8
ENSMUST00000197385.2 |
Cdk6
|
cyclin-dependent kinase 6 |
| chr2_-_174188505 | 0.72 |
ENSMUST00000168292.2
|
Gm20721
|
predicted gene, 20721 |
| chr9_+_106158212 | 0.69 |
ENSMUST00000072206.14
|
Poc1a
|
POC1 centriolar protein A |
| chr1_-_182929025 | 0.67 |
ENSMUST00000171366.7
|
Disp1
|
dispatched RND transporter family member 1 |
| chr15_-_91075933 | 0.61 |
ENSMUST00000069511.8
|
Abcd2
|
ATP-binding cassette, sub-family D (ALD), member 2 |
| chrX_+_158491589 | 0.60 |
ENSMUST00000080394.13
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr13_+_37529184 | 0.59 |
ENSMUST00000021860.7
|
Ly86
|
lymphocyte antigen 86 |
| chr1_+_150268544 | 0.59 |
ENSMUST00000124973.9
|
Tpr
|
translocated promoter region, nuclear basket protein |
| chr19_+_37184927 | 0.58 |
ENSMUST00000024078.15
ENSMUST00000112391.8 |
Marchf5
|
membrane associated ring-CH-type finger 5 |
| chr5_-_74229021 | 0.57 |
ENSMUST00000119154.8
ENSMUST00000068058.14 |
Usp46
|
ubiquitin specific peptidase 46 |
| chr19_-_37153436 | 0.56 |
ENSMUST00000142973.2
ENSMUST00000154376.8 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr2_+_173918715 | 0.55 |
ENSMUST00000087908.10
ENSMUST00000044638.13 ENSMUST00000156054.2 |
Stx16
|
syntaxin 16 |
| chr18_+_34892599 | 0.55 |
ENSMUST00000097622.4
|
Fam53c
|
family with sequence similarity 53, member C |
| chr19_-_53360197 | 0.52 |
ENSMUST00000086887.2
|
Gm10197
|
predicted gene 10197 |
| chr6_+_5390386 | 0.52 |
ENSMUST00000183358.2
|
Asb4
|
ankyrin repeat and SOCS box-containing 4 |
| chr15_+_84553801 | 0.51 |
ENSMUST00000171460.8
|
Prr5
|
proline rich 5 (renal) |
| chr11_+_120123727 | 0.49 |
ENSMUST00000122148.8
ENSMUST00000044985.14 |
Bahcc1
|
BAH domain and coiled-coil containing 1 |
| chr11_-_69811347 | 0.49 |
ENSMUST00000108610.8
|
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr11_-_115494692 | 0.48 |
ENSMUST00000125097.2
ENSMUST00000019135.14 ENSMUST00000106508.10 |
Gga3
|
golgi associated, gamma adaptin ear containing, ARF binding protein 3 |
| chr11_+_100211363 | 0.45 |
ENSMUST00000152521.2
|
Eif1
|
eukaryotic translation initiation factor 1 |
| chr11_-_87295292 | 0.45 |
ENSMUST00000067692.13
|
Rad51c
|
RAD51 paralog C |
| chr3_-_59127571 | 0.44 |
ENSMUST00000199675.2
ENSMUST00000170388.6 |
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr11_-_101308441 | 0.42 |
ENSMUST00000070395.9
|
Aarsd1
|
alanyl-tRNA synthetase domain containing 1 |
| chr1_+_177272215 | 0.42 |
ENSMUST00000192851.2
ENSMUST00000193480.2 ENSMUST00000195388.2 |
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr1_-_170133901 | 0.41 |
ENSMUST00000179801.3
|
Gm7694
|
predicted gene 7694 |
| chr5_-_90487583 | 0.41 |
ENSMUST00000197021.2
|
Ankrd17
|
ankyrin repeat domain 17 |
| chr3_-_95902949 | 0.40 |
ENSMUST00000123006.8
ENSMUST00000130043.8 |
Plekho1
|
pleckstrin homology domain containing, family O member 1 |
| chr13_+_108182941 | 0.39 |
ENSMUST00000224361.2
ENSMUST00000224390.2 |
Smim15
|
small integral membrane protein 15 |
| chrX_+_35592006 | 0.38 |
ENSMUST00000016383.10
|
Lonrf3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr3_-_129834788 | 0.38 |
ENSMUST00000168644.3
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae) |
| chr11_+_95734028 | 0.36 |
ENSMUST00000107709.8
|
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr9_+_111011327 | 0.33 |
ENSMUST00000216430.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr2_-_71198091 | 0.32 |
ENSMUST00000151937.8
|
Slc25a12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12 |
| chr17_-_47143214 | 0.32 |
ENSMUST00000233537.2
|
Bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr5_+_75312939 | 0.31 |
ENSMUST00000202681.4
ENSMUST00000000476.15 |
Pdgfra
|
platelet derived growth factor receptor, alpha polypeptide |
| chr9_+_111011388 | 0.31 |
ENSMUST00000217117.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr16_+_57173632 | 0.30 |
ENSMUST00000099667.3
|
Filip1l
|
filamin A interacting protein 1-like |
| chr10_+_70040504 | 0.28 |
ENSMUST00000190199.2
|
Mrln
|
myoregulin |
| chr12_-_114901026 | 0.27 |
ENSMUST00000103516.2
ENSMUST00000191868.2 |
Ighv1-42
|
immunoglobulin heavy variable V1-42 |
| chr11_+_95734419 | 0.27 |
ENSMUST00000107708.2
|
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr10_+_70040483 | 0.26 |
ENSMUST00000020090.8
|
Mrln
|
myoregulin |
| chr7_+_4928784 | 0.26 |
ENSMUST00000057612.9
|
Ssc5d
|
scavenger receptor cysteine rich family, 5 domains |
| chr5_-_120887864 | 0.26 |
ENSMUST00000053909.13
ENSMUST00000081491.13 |
Oas2
|
2'-5' oligoadenylate synthetase 2 |
| chr11_+_68393845 | 0.24 |
ENSMUST00000102613.8
ENSMUST00000060441.7 |
Pik3r6
|
phosphoinositide-3-kinase regulatory subunit 5 |
| chr9_+_19620729 | 0.23 |
ENSMUST00000217450.4
ENSMUST00000212013.4 |
Olfr857
|
olfactory receptor 857 |
| chr6_-_128252540 | 0.22 |
ENSMUST00000130454.8
|
Tead4
|
TEA domain family member 4 |
| chr15_+_101071948 | 0.21 |
ENSMUST00000000544.12
|
Acvr1b
|
activin A receptor, type 1B |
| chr7_+_30399208 | 0.21 |
ENSMUST00000013227.8
|
2200002J24Rik
|
RIKEN cDNA 2200002J24 gene |
| chr5_-_124325213 | 0.20 |
ENSMUST00000161938.8
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr17_-_48189815 | 0.20 |
ENSMUST00000154108.2
|
Foxp4
|
forkhead box P4 |
| chr9_+_19716202 | 0.19 |
ENSMUST00000212540.3
ENSMUST00000217280.2 |
Olfr859
|
olfactory receptor 859 |
| chr6_-_12749192 | 0.19 |
ENSMUST00000172356.8
|
Thsd7a
|
thrombospondin, type I, domain containing 7A |
| chr8_+_26092252 | 0.19 |
ENSMUST00000136107.9
ENSMUST00000146919.8 ENSMUST00000139966.8 ENSMUST00000142395.8 ENSMUST00000143445.2 |
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr5_+_123648523 | 0.17 |
ENSMUST00000031384.6
|
B3gnt4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr1_+_177273226 | 0.17 |
ENSMUST00000077225.8
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr4_-_82768958 | 0.16 |
ENSMUST00000139401.2
|
Zdhhc21
|
zinc finger, DHHC domain containing 21 |
| chr17_-_87255295 | 0.15 |
ENSMUST00000233995.2
ENSMUST00000065758.8 ENSMUST00000235110.2 |
Atp6v1e2
|
ATPase, H+ transporting, lysosomal V1 subunit E2 |
| chr1_+_156386414 | 0.15 |
ENSMUST00000166172.9
ENSMUST00000027888.13 |
Abl2
|
v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene) |
| chr3_-_123029782 | 0.15 |
ENSMUST00000106427.8
ENSMUST00000198584.2 |
Synpo2
|
synaptopodin 2 |
| chr3_+_137770813 | 0.14 |
ENSMUST00000163080.3
|
1110002E22Rik
|
RIKEN cDNA 1110002E22 gene |
| chr12_+_86781154 | 0.14 |
ENSMUST00000095527.6
|
Lrrc74a
|
leucine rich repeat containing 74A |
| chr2_+_61876806 | 0.14 |
ENSMUST00000102735.10
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 |
| chr2_-_25471703 | 0.14 |
ENSMUST00000114217.3
ENSMUST00000191602.2 |
Ajm1
|
apical junction component 1 |
| chr13_+_16189041 | 0.14 |
ENSMUST00000164993.2
|
Inhba
|
inhibin beta-A |
| chr7_+_17799863 | 0.13 |
ENSMUST00000108487.9
|
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chr15_-_89726063 | 0.13 |
ENSMUST00000029441.4
|
Syt10
|
synaptotagmin X |
| chr5_-_142594549 | 0.13 |
ENSMUST00000037048.9
|
Mmd2
|
monocyte to macrophage differentiation-associated 2 |
| chr15_+_99870787 | 0.13 |
ENSMUST00000231160.2
|
Larp4
|
La ribonucleoprotein domain family, member 4 |
| chr4_-_110149916 | 0.13 |
ENSMUST00000106601.8
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr1_-_131441962 | 0.12 |
ENSMUST00000185445.3
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr3_+_125197722 | 0.12 |
ENSMUST00000173932.8
|
Ndst4
|
N-deacetylase/N-sulfotransferase (heparin glucosaminyl) 4 |
| chr9_+_26910995 | 0.11 |
ENSMUST00000213770.2
ENSMUST00000213683.2 ENSMUST00000039161.10 |
Thyn1
|
thymocyte nuclear protein 1 |
| chr7_+_17799849 | 0.11 |
ENSMUST00000032520.9
|
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chr7_+_17799889 | 0.11 |
ENSMUST00000108483.2
|
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chr6_-_52203146 | 0.10 |
ENSMUST00000114425.3
|
Hoxa9
|
homeobox A9 |
| chr6_-_12749409 | 0.10 |
ENSMUST00000119581.7
|
Thsd7a
|
thrombospondin, type I, domain containing 7A |
| chr18_+_74349189 | 0.10 |
ENSMUST00000025444.8
|
Cxxc1
|
CXXC finger 1 (PHD domain) |
| chr8_+_11778039 | 0.10 |
ENSMUST00000110909.9
|
Arhgef7
|
Rho guanine nucleotide exchange factor (GEF7) |
| chr6_+_17636979 | 0.09 |
ENSMUST00000015877.14
ENSMUST00000152005.3 |
Capza2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr2_-_79287095 | 0.09 |
ENSMUST00000041099.5
|
Neurod1
|
neurogenic differentiation 1 |
| chr7_-_119461027 | 0.09 |
ENSMUST00000137888.2
ENSMUST00000142120.2 |
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr16_-_17540685 | 0.09 |
ENSMUST00000232163.2
ENSMUST00000232202.2 ENSMUST00000080936.14 ENSMUST00000232645.2 ENSMUST00000232431.2 |
Med15
|
mediator complex subunit 15 |
| chr2_-_36782948 | 0.09 |
ENSMUST00000217215.3
|
Olfr353
|
olfactory receptor 353 |
| chr11_+_53324126 | 0.08 |
ENSMUST00000018382.7
|
Gdf9
|
growth differentiation factor 9 |
| chr7_-_133203838 | 0.08 |
ENSMUST00000033275.4
|
Tex36
|
testis expressed 36 |
| chr16_-_26810402 | 0.08 |
ENSMUST00000231299.2
|
Gmnc
|
geminin coiled-coil domain containing |
| chr11_+_73051228 | 0.07 |
ENSMUST00000006104.10
ENSMUST00000135202.8 ENSMUST00000136894.3 |
P2rx5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr3_-_10366229 | 0.06 |
ENSMUST00000119761.2
ENSMUST00000029043.13 |
Fabp12
|
fatty acid binding protein 12 |
| chr11_-_69811717 | 0.05 |
ENSMUST00000152589.2
ENSMUST00000108612.8 ENSMUST00000108611.8 |
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr4_+_136150835 | 0.05 |
ENSMUST00000088677.5
ENSMUST00000121571.8 ENSMUST00000117699.2 |
Htr1d
|
5-hydroxytryptamine (serotonin) receptor 1D |
| chr6_-_3494587 | 0.05 |
ENSMUST00000049985.15
|
Hepacam2
|
HEPACAM family member 2 |
| chr6_-_124208815 | 0.05 |
ENSMUST00000233936.2
ENSMUST00000100968.4 |
Vmn2r27
|
vomeronasal 2, receptor27 |
| chr7_+_55443931 | 0.05 |
ENSMUST00000205796.2
|
Tubgcp5
|
tubulin, gamma complex associated protein 5 |
| chr7_+_55443893 | 0.05 |
ENSMUST00000032627.5
|
Tubgcp5
|
tubulin, gamma complex associated protein 5 |
| chr2_-_156022054 | 0.04 |
ENSMUST00000126992.8
ENSMUST00000146288.8 ENSMUST00000029149.13 ENSMUST00000109587.9 ENSMUST00000109584.8 |
Rbm39
|
RNA binding motif protein 39 |
| chr16_-_17540805 | 0.04 |
ENSMUST00000012259.9
ENSMUST00000232236.2 |
Med15
|
mediator complex subunit 15 |
| chr16_+_43056218 | 0.04 |
ENSMUST00000146708.8
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr12_+_86781141 | 0.03 |
ENSMUST00000223308.2
|
Lrrc74a
|
leucine rich repeat containing 74A |
| chr11_+_73244561 | 0.03 |
ENSMUST00000108465.4
|
Olfr20
|
olfactory receptor 20 |
| chr14_-_79461316 | 0.03 |
ENSMUST00000040802.5
|
Zfp957
|
zinc finger protein 957 |
| chr2_+_111491764 | 0.03 |
ENSMUST00000213511.2
ENSMUST00000207228.3 ENSMUST00000208983.2 |
Olfr1299
Gm44840
|
olfactory receptor 1299 predicted gene 44840 |
| chr11_+_117157024 | 0.02 |
ENSMUST00000019038.15
|
Septin9
|
septin 9 |
| chr11_+_49138278 | 0.02 |
ENSMUST00000109194.2
|
Mgat1
|
mannoside acetylglucosaminyltransferase 1 |
| chr11_-_76468527 | 0.02 |
ENSMUST00000176179.8
|
Abr
|
active BCR-related gene |
| chr6_-_113478779 | 0.02 |
ENSMUST00000101059.4
ENSMUST00000204268.3 ENSMUST00000205170.2 ENSMUST00000205075.2 ENSMUST00000204134.3 |
Prrt3
|
proline-rich transmembrane protein 3 |
| chr15_-_27788693 | 0.02 |
ENSMUST00000226287.2
|
Trio
|
triple functional domain (PTPRF interacting) |
| chr1_+_9978863 | 0.01 |
ENSMUST00000052843.12
ENSMUST00000171802.8 ENSMUST00000125294.9 ENSMUST00000140948.9 |
Mcmdc2
|
minichromosome maintenance domain containing 2 |
| chr16_-_59090028 | 0.01 |
ENSMUST00000099656.2
|
Olfr201
|
olfactory receptor 201 |
| chr6_+_91881193 | 0.01 |
ENSMUST00000205686.2
|
4930590J08Rik
|
RIKEN cDNA 4930590J08 gene |
| chr15_+_101013704 | 0.01 |
ENSMUST00000229954.2
|
Ankrd33
|
ankyrin repeat domain 33 |
| chr19_-_11833365 | 0.01 |
ENSMUST00000079875.4
|
Olfr1418
|
olfactory receptor 1418 |
| chr15_+_101013619 | 0.00 |
ENSMUST00000070875.8
ENSMUST00000231158.2 |
Ankrd33
|
ankyrin repeat domain 33 |
| chr16_-_57113207 | 0.00 |
ENSMUST00000023434.15
ENSMUST00000120112.2 ENSMUST00000119407.8 |
Tmem30c
|
transmembrane protein 30C |
| chr3_-_107240989 | 0.00 |
ENSMUST00000061772.11
|
Rbm15
|
RNA binding motif protein 15 |
| chr2_-_38955452 | 0.00 |
ENSMUST00000112850.9
|
Golga1
|
golgi autoantigen, golgin subfamily a, 1 |
| chr15_+_99870714 | 0.00 |
ENSMUST00000230956.2
|
Larp4
|
La ribonucleoprotein domain family, member 4 |
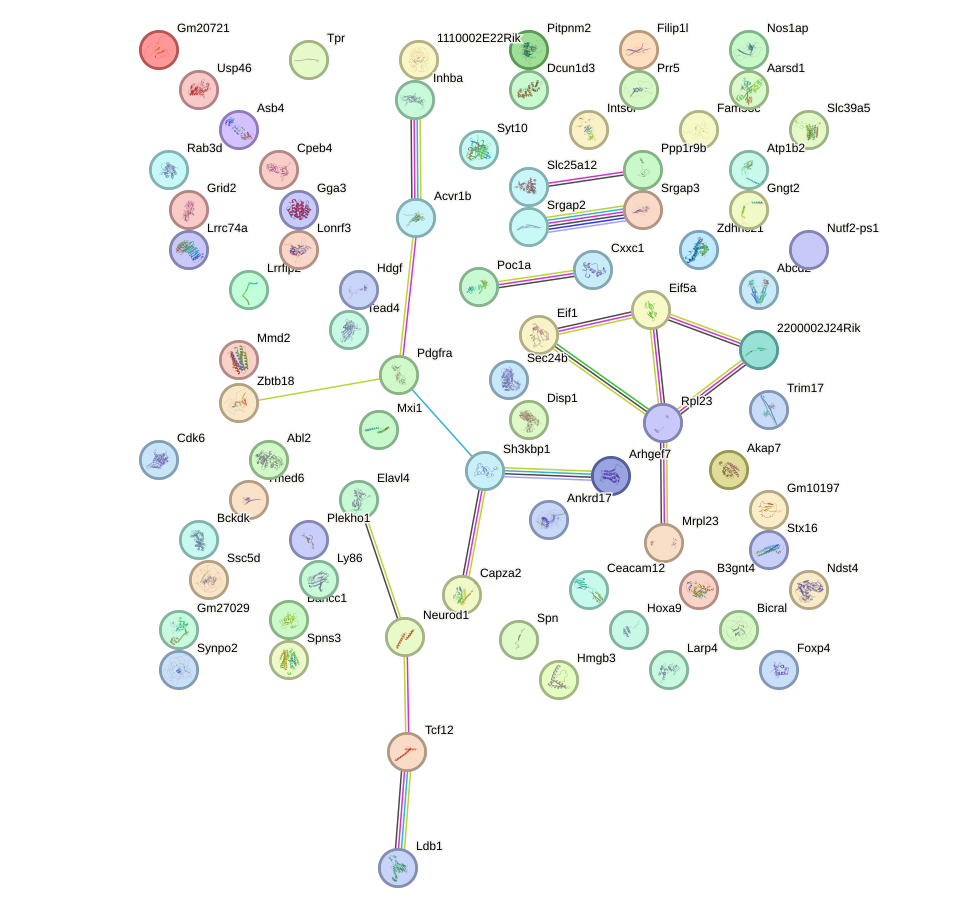
Network of associatons between targets according to the STRING database.
First level regulatory network of Atoh1_Bhlhe23
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.7 | 5.4 | GO:0032796 | uropod organization(GO:0032796) early endosome to recycling endosome transport(GO:0061502) |
| 0.6 | 1.8 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) |
| 0.4 | 1.1 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.3 | 1.7 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.3 | 1.7 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.3 | 1.3 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.2 | 0.7 | GO:0036275 | response to 5-fluorouracil(GO:0036275) |
| 0.2 | 0.7 | GO:0007225 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.2 | 1.3 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.2 | 0.6 | GO:0046832 | RNA import into nucleus(GO:0006404) mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.2 | 0.6 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.2 | 0.8 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.1 | 0.9 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.4 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 3.0 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 0.5 | GO:1902080 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.1 | 1.1 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.6 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.1 | 0.9 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 0.4 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.1 | 0.8 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.4 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 0.5 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.4 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.1 | 0.7 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 | 0.3 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 0.4 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.3 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.6 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 0.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.3 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.0 | 0.6 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.2 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.0 | 0.2 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.5 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.2 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.5 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.6 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 3.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.3 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.0 | 0.5 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.1 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.6 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.4 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.2 | GO:1901250 | regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.0 | 0.1 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.0 | 0.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.0 | 0.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.9 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.2 | 0.7 | GO:0097132 | cyclin D2-CDK6 complex(GO:0097132) |
| 0.2 | 5.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.2 | 1.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 1.8 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.4 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.6 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.4 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 1.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.0 | 0.5 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.5 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 1.7 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 3.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.9 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.3 | 5.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 0.7 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.2 | 1.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.9 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 0.4 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 1.7 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 5.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.3 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 1.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 1.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.8 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.4 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 1.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.7 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.5 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.9 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.9 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.6 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 1.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.3 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 3.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 1.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.4 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |