Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
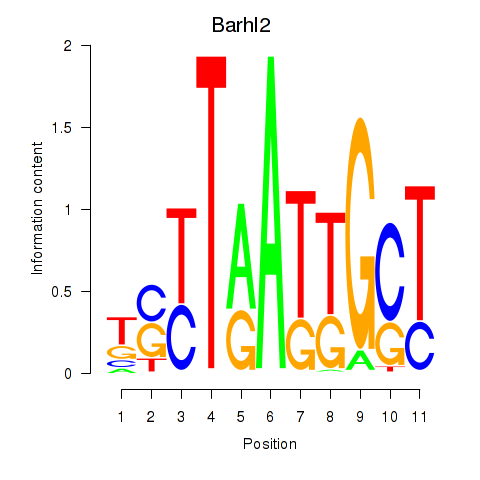
Results for Barhl2
Z-value: 0.39
Transcription factors associated with Barhl2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Barhl2
|
ENSMUSG00000034384.12 | BarH like homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Barhl2 | mm39_v1_chr5_-_106606032_106606032 | -0.36 | 3.1e-02 | Click! |
Activity profile of Barhl2 motif
Sorted Z-values of Barhl2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_110987604 | 0.99 |
ENSMUST00000056937.12
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr5_-_110987441 | 0.97 |
ENSMUST00000145318.2
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr7_+_123061497 | 0.73 |
ENSMUST00000033023.10
|
Aqp8
|
aquaporin 8 |
| chr7_+_123061535 | 0.72 |
ENSMUST00000098056.6
|
Aqp8
|
aquaporin 8 |
| chr11_-_95733235 | 0.68 |
ENSMUST00000059026.10
|
Abi3
|
ABI family member 3 |
| chr2_-_25114660 | 0.59 |
ENSMUST00000043584.5
|
Tubb4b
|
tubulin, beta 4B class IVB |
| chr10_+_127734384 | 0.58 |
ENSMUST00000047134.8
|
Sdr9c7
|
4short chain dehydrogenase/reductase family 9C, member 7 |
| chr7_-_48497771 | 0.56 |
ENSMUST00000032658.14
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr15_+_76579885 | 0.50 |
ENSMUST00000231028.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr7_+_24069680 | 0.49 |
ENSMUST00000205428.2
ENSMUST00000171904.3 ENSMUST00000205626.2 |
Kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr7_+_110368037 | 0.47 |
ENSMUST00000213373.2
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr5_+_110987839 | 0.45 |
ENSMUST00000200172.2
ENSMUST00000066160.3 |
Chek2
|
checkpoint kinase 2 |
| chr7_+_126810780 | 0.43 |
ENSMUST00000032910.13
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr14_+_36776775 | 0.43 |
ENSMUST00000120052.2
|
Lrit1
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1 |
| chr15_+_76579960 | 0.38 |
ENSMUST00000229679.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr16_+_38182569 | 0.37 |
ENSMUST00000023494.13
ENSMUST00000114739.2 |
Popdc2
|
popeye domain containing 2 |
| chr2_+_143757193 | 0.36 |
ENSMUST00000103172.4
|
Dstn
|
destrin |
| chr18_-_68562385 | 0.36 |
ENSMUST00000052347.8
|
Mc2r
|
melanocortin 2 receptor |
| chr1_+_91178017 | 0.33 |
ENSMUST00000059743.12
ENSMUST00000178627.8 ENSMUST00000171165.8 ENSMUST00000191368.7 |
Ube2f
|
ubiquitin-conjugating enzyme E2F (putative) |
| chr1_+_91178288 | 0.32 |
ENSMUST00000171112.8
ENSMUST00000191533.2 |
Ube2f
|
ubiquitin-conjugating enzyme E2F (putative) |
| chr7_+_28580847 | 0.31 |
ENSMUST00000066880.6
|
Capn12
|
calpain 12 |
| chr2_-_3513783 | 0.30 |
ENSMUST00000124331.8
ENSMUST00000140494.2 ENSMUST00000027961.12 |
Gm45902
Hspa14
|
predicted gene 45902 heat shock protein 14 |
| chr5_-_115236354 | 0.27 |
ENSMUST00000100848.3
|
Gm10401
|
predicted gene 10401 |
| chrX_+_7688528 | 0.26 |
ENSMUST00000009875.5
|
Kcnd1
|
potassium voltage-gated channel, Shal-related family, member 1 |
| chr8_-_84321032 | 0.25 |
ENSMUST00000163837.2
|
Tecr
|
trans-2,3-enoyl-CoA reductase |
| chr5_+_120727068 | 0.25 |
ENSMUST00000069259.9
ENSMUST00000094391.6 |
Iqcd
|
IQ motif containing D |
| chr8_+_95807814 | 0.24 |
ENSMUST00000034239.9
|
Katnb1
|
katanin p80 (WD40-containing) subunit B 1 |
| chr8_+_46463633 | 0.24 |
ENSMUST00000110381.9
|
Lrp2bp
|
Lrp2 binding protein |
| chr10_-_99595498 | 0.23 |
ENSMUST00000056085.6
|
Csl
|
citrate synthase like |
| chr17_-_25946370 | 0.23 |
ENSMUST00000170070.3
ENSMUST00000048054.14 |
Chtf18
|
CTF18, chromosome transmission fidelity factor 18 |
| chr8_+_18896267 | 0.23 |
ENSMUST00000149565.8
ENSMUST00000033847.5 |
Agpat5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon) |
| chr5_+_24679154 | 0.23 |
ENSMUST00000199856.2
|
Agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr11_+_116324913 | 0.22 |
ENSMUST00000057676.7
|
Ubald2
|
UBA-like domain containing 2 |
| chr11_+_49327451 | 0.22 |
ENSMUST00000215226.2
|
Olfr1388
|
olfactory receptor 1388 |
| chr2_-_129541753 | 0.22 |
ENSMUST00000028883.12
|
Pdyn
|
prodynorphin |
| chr2_+_9887427 | 0.22 |
ENSMUST00000114919.2
|
4930412O13Rik
|
RIKEN cDNA 4930412O13 gene |
| chr7_+_3632982 | 0.21 |
ENSMUST00000179769.8
ENSMUST00000008517.13 |
Prpf31
|
pre-mRNA processing factor 31 |
| chr12_-_108145498 | 0.21 |
ENSMUST00000071095.14
|
Setd3
|
SET domain containing 3 |
| chr17_-_27158514 | 0.21 |
ENSMUST00000114935.9
ENSMUST00000025027.10 |
Cuta
|
cutA divalent cation tolerance homolog |
| chr12_+_72488625 | 0.20 |
ENSMUST00000161284.3
ENSMUST00000162159.8 |
Lrrc9
|
leucine rich repeat containing 9 |
| chr8_-_84321069 | 0.20 |
ENSMUST00000019382.17
ENSMUST00000212630.2 ENSMUST00000165740.9 |
Tecr
|
trans-2,3-enoyl-CoA reductase |
| chr16_-_36648586 | 0.20 |
ENSMUST00000023537.6
ENSMUST00000114829.9 |
Eaf2
|
ELL associated factor 2 |
| chr4_+_19818718 | 0.20 |
ENSMUST00000035890.8
|
Slc7a13
|
solute carrier family 7, (cationic amino acid transporter, y+ system) member 13 |
| chr11_+_62711295 | 0.19 |
ENSMUST00000108703.2
|
Trim16
|
tripartite motif-containing 16 |
| chr4_-_49597425 | 0.19 |
ENSMUST00000150664.2
|
Pgap4
|
post-GPI attachment to proteins GalNAc transferase 4 |
| chr19_+_24853039 | 0.19 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr11_-_40646090 | 0.19 |
ENSMUST00000020576.8
|
Ccng1
|
cyclin G1 |
| chr12_+_108145802 | 0.19 |
ENSMUST00000221167.2
|
Ccnk
|
cyclin K |
| chr14_+_53698556 | 0.19 |
ENSMUST00000181728.3
|
Trav7-4
|
T cell receptor alpha variable 7-4 |
| chr11_-_100098333 | 0.19 |
ENSMUST00000007272.8
|
Krt14
|
keratin 14 |
| chr9_-_13738204 | 0.19 |
ENSMUST00000147115.8
|
Cep57
|
centrosomal protein 57 |
| chr2_-_31700592 | 0.18 |
ENSMUST00000057407.3
|
Qrfp
|
pyroglutamylated RFamide peptide |
| chr6_+_57234937 | 0.18 |
ENSMUST00000228297.2
|
Vmn1r15
|
vomeronasal 1 receptor 15 |
| chr11_+_62711057 | 0.18 |
ENSMUST00000055006.12
ENSMUST00000072639.10 |
Trim16
|
tripartite motif-containing 16 |
| chr2_+_57887896 | 0.18 |
ENSMUST00000112616.8
ENSMUST00000166729.2 |
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr14_-_30850795 | 0.18 |
ENSMUST00000049732.11
ENSMUST00000090205.5 ENSMUST00000064032.10 |
Smim4
|
small integral membrane protein 4 |
| chr3_-_80710097 | 0.18 |
ENSMUST00000075316.10
ENSMUST00000107745.8 |
Gria2
|
glutamate receptor, ionotropic, AMPA2 (alpha 2) |
| chr2_-_103114105 | 0.17 |
ENSMUST00000111174.8
|
Ehf
|
ets homologous factor |
| chr1_+_165616315 | 0.17 |
ENSMUST00000161559.3
|
Cd247
|
CD247 antigen |
| chr4_-_141125885 | 0.17 |
ENSMUST00000133676.3
ENSMUST00000042617.14 |
Clcnka
|
chloride channel, voltage-sensitive Ka |
| chr11_-_116743898 | 0.17 |
ENSMUST00000190993.7
ENSMUST00000092404.13 |
Srsf2
|
serine and arginine-rich splicing factor 2 |
| chr10_+_73657753 | 0.17 |
ENSMUST00000134009.9
ENSMUST00000177420.7 ENSMUST00000125006.9 |
Pcdh15
|
protocadherin 15 |
| chr7_-_118304930 | 0.17 |
ENSMUST00000207323.2
ENSMUST00000038791.15 |
Gde1
|
glycerophosphodiester phosphodiesterase 1 |
| chr14_+_21550921 | 0.17 |
ENSMUST00000182964.3
|
Kat6b
|
K(lysine) acetyltransferase 6B |
| chr4_-_120427449 | 0.16 |
ENSMUST00000030381.8
|
Ctps
|
cytidine 5'-triphosphate synthase |
| chr15_-_100321973 | 0.16 |
ENSMUST00000154676.2
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr2_+_3514071 | 0.16 |
ENSMUST00000036350.3
|
Cdnf
|
cerebral dopamine neurotrophic factor |
| chr8_+_10299288 | 0.16 |
ENSMUST00000214643.2
|
Myo16
|
myosin XVI |
| chr7_-_19449319 | 0.16 |
ENSMUST00000032555.10
ENSMUST00000093552.12 |
Tomm40
|
translocase of outer mitochondrial membrane 40 |
| chr4_-_141143313 | 0.16 |
ENSMUST00000006378.9
ENSMUST00000105788.2 |
Clcnkb
|
chloride channel, voltage-sensitive Kb |
| chr14_+_52892115 | 0.16 |
ENSMUST00000198019.2
|
Trav7-1
|
T cell receptor alpha variable 7-1 |
| chrX_+_70707271 | 0.16 |
ENSMUST00000070449.6
|
Gpr50
|
G-protein-coupled receptor 50 |
| chr16_-_9812787 | 0.16 |
ENSMUST00000199708.5
|
Grin2a
|
glutamate receptor, ionotropic, NMDA2A (epsilon 1) |
| chr2_-_181101158 | 0.16 |
ENSMUST00000155535.2
ENSMUST00000029106.13 ENSMUST00000087409.10 |
Zbtb46
|
zinc finger and BTB domain containing 46 |
| chr12_+_108145997 | 0.16 |
ENSMUST00000101055.5
|
Ccnk
|
cyclin K |
| chr5_+_20112500 | 0.16 |
ENSMUST00000101558.10
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr7_+_79309938 | 0.16 |
ENSMUST00000035977.9
|
Ticrr
|
TOPBP1-interacting checkpoint and replication regulator |
| chr12_-_104439589 | 0.15 |
ENSMUST00000021513.6
|
Gsc
|
goosecoid homeobox |
| chr14_-_68771138 | 0.15 |
ENSMUST00000022640.8
|
Adam7
|
a disintegrin and metallopeptidase domain 7 |
| chr18_+_34869412 | 0.15 |
ENSMUST00000105038.3
|
Gm3550
|
predicted gene 3550 |
| chr5_-_123663440 | 0.15 |
ENSMUST00000197682.6
|
Gm49027
|
predicted gene, 49027 |
| chr1_+_173983199 | 0.15 |
ENSMUST00000213748.2
|
Olfr420
|
olfactory receptor 420 |
| chr14_-_88708782 | 0.15 |
ENSMUST00000192557.2
ENSMUST00000061628.7 |
Pcdh20
|
protocadherin 20 |
| chr3_+_95496239 | 0.15 |
ENSMUST00000177390.8
ENSMUST00000060323.12 ENSMUST00000098861.11 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr15_+_66542598 | 0.15 |
ENSMUST00000065916.14
|
Tg
|
thyroglobulin |
| chr17_+_25946644 | 0.15 |
ENSMUST00000237183.2
ENSMUST00000237785.2 ENSMUST00000047273.3 |
Rpusd1
|
RNA pseudouridylate synthase domain containing 1 |
| chr13_+_118851214 | 0.15 |
ENSMUST00000022246.9
|
Fgf10
|
fibroblast growth factor 10 |
| chr2_+_125876883 | 0.15 |
ENSMUST00000110442.2
|
Fgf7
|
fibroblast growth factor 7 |
| chr7_-_12063408 | 0.14 |
ENSMUST00000226701.2
|
Vmn1r83
|
vomeronasal 1 receptor 83 |
| chr1_-_163552693 | 0.14 |
ENSMUST00000159679.8
|
Mettl11b
|
methyltransferase like 11B |
| chr2_+_155850002 | 0.14 |
ENSMUST00000088650.11
|
Ergic3
|
ERGIC and golgi 3 |
| chr1_-_14380032 | 0.14 |
ENSMUST00000187790.2
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr1_-_67077906 | 0.14 |
ENSMUST00000119559.8
ENSMUST00000149996.2 ENSMUST00000027149.12 ENSMUST00000113979.10 |
Lancl1
|
LanC (bacterial lantibiotic synthetase component C)-like 1 |
| chr18_+_4993795 | 0.14 |
ENSMUST00000153016.8
|
Svil
|
supervillin |
| chr17_+_34544632 | 0.14 |
ENSMUST00000050325.10
|
H2-Eb2
|
histocompatibility 2, class II antigen E beta2 |
| chr8_-_62576140 | 0.14 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr14_-_21764638 | 0.13 |
ENSMUST00000073870.7
|
Dupd1
|
dual specificity phosphatase and pro isomerase domain containing 1 |
| chr15_-_34356567 | 0.13 |
ENSMUST00000179647.2
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr6_-_116561156 | 0.13 |
ENSMUST00000061723.6
|
Olfr215
|
olfactory receptor 215 |
| chr7_+_100145192 | 0.13 |
ENSMUST00000133044.3
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr6_-_8259098 | 0.13 |
ENSMUST00000012627.5
|
Rpa3
|
replication protein A3 |
| chr3_+_95496270 | 0.13 |
ENSMUST00000176674.8
ENSMUST00000177389.8 ENSMUST00000176755.8 ENSMUST00000177399.2 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr2_+_155849965 | 0.13 |
ENSMUST00000006035.13
|
Ergic3
|
ERGIC and golgi 3 |
| chr1_-_153775944 | 0.13 |
ENSMUST00000123490.2
|
Teddm2
|
transmembrane epididymal family member 2 |
| chr2_+_31530049 | 0.13 |
ENSMUST00000113470.3
|
Prdm12
|
PR domain containing 12 |
| chr10_-_89093441 | 0.13 |
ENSMUST00000182341.8
ENSMUST00000182613.8 |
Ano4
|
anoctamin 4 |
| chr15_+_98972850 | 0.12 |
ENSMUST00000039665.8
|
Troap
|
trophinin associated protein |
| chr2_+_125876566 | 0.12 |
ENSMUST00000064794.14
|
Fgf7
|
fibroblast growth factor 7 |
| chr16_-_9812410 | 0.12 |
ENSMUST00000115835.8
|
Grin2a
|
glutamate receptor, ionotropic, NMDA2A (epsilon 1) |
| chr16_+_10652910 | 0.12 |
ENSMUST00000037913.9
|
Rmi2
|
RecQ mediated genome instability 2 |
| chr5_-_123185029 | 0.12 |
ENSMUST00000045843.15
|
Morn3
|
MORN repeat containing 3 |
| chr5_-_65855511 | 0.12 |
ENSMUST00000201948.4
|
Pds5a
|
PDS5 cohesin associated factor A |
| chr17_+_35439810 | 0.12 |
ENSMUST00000118793.2
|
Gm16181
|
predicted gene 16181 |
| chr5_-_86824205 | 0.12 |
ENSMUST00000038448.7
|
Tmprss11b
|
transmembrane protease, serine 11B |
| chr18_+_61820982 | 0.11 |
ENSMUST00000025471.4
|
Il17b
|
interleukin 17B |
| chr14_+_53007210 | 0.11 |
ENSMUST00000178768.4
|
Trav7d-4
|
T cell receptor alpha variable 7D-4 |
| chrX_-_156198282 | 0.11 |
ENSMUST00000079945.11
ENSMUST00000138396.3 |
Phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr15_-_101482320 | 0.11 |
ENSMUST00000042957.6
|
Krt75
|
keratin 75 |
| chr15_-_82872073 | 0.11 |
ENSMUST00000229439.2
|
Tcf20
|
transcription factor 20 |
| chr10_+_73657689 | 0.11 |
ENSMUST00000064562.14
ENSMUST00000193174.6 ENSMUST00000105426.10 ENSMUST00000129404.9 ENSMUST00000131321.9 ENSMUST00000126920.9 ENSMUST00000147189.9 ENSMUST00000105424.10 ENSMUST00000092420.13 ENSMUST00000105429.10 ENSMUST00000193361.6 ENSMUST00000131724.9 ENSMUST00000152655.9 ENSMUST00000151116.9 ENSMUST00000155701.9 ENSMUST00000152819.9 ENSMUST00000125517.9 ENSMUST00000124046.8 ENSMUST00000146682.8 ENSMUST00000177107.8 ENSMUST00000149977.9 ENSMUST00000191854.6 |
Pcdh15
|
protocadherin 15 |
| chr1_+_153767478 | 0.11 |
ENSMUST00000050660.6
|
Teddm1a
|
transmembrane epididymal protein 1A |
| chr3_-_95158457 | 0.11 |
ENSMUST00000125515.3
ENSMUST00000107195.9 |
Bnipl
|
BCL2/adenovirus E1B 19kD interacting protein like |
| chr19_-_13828056 | 0.11 |
ENSMUST00000208493.3
|
Olfr1501
|
olfactory receptor 1501 |
| chr17_-_37472385 | 0.10 |
ENSMUST00000219235.3
|
Olfr93
|
olfactory receptor 93 |
| chr2_-_164670452 | 0.10 |
ENSMUST00000017911.4
|
Spata25
|
spermatogenesis associated 25 |
| chr9_-_13738304 | 0.10 |
ENSMUST00000148086.8
ENSMUST00000034398.12 |
Cep57
|
centrosomal protein 57 |
| chr5_-_123185073 | 0.10 |
ENSMUST00000031437.14
|
Morn3
|
MORN repeat containing 3 |
| chr19_+_13208692 | 0.10 |
ENSMUST00000207246.4
|
Olfr1463
|
olfactory receptor 1463 |
| chr7_-_3632826 | 0.10 |
ENSMUST00000205596.2
ENSMUST00000155592.8 ENSMUST00000108641.10 |
Tfpt
|
TCF3 (E2A) fusion partner |
| chr5_+_20112704 | 0.10 |
ENSMUST00000115267.7
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr2_-_126975804 | 0.10 |
ENSMUST00000110387.4
|
Ncaph
|
non-SMC condensin I complex, subunit H |
| chr5_+_110988095 | 0.10 |
ENSMUST00000198373.2
|
Chek2
|
checkpoint kinase 2 |
| chr11_+_60308077 | 0.10 |
ENSMUST00000070681.7
|
Gid4
|
GID complex subunit 4, VID24 homolog |
| chr7_+_12034357 | 0.10 |
ENSMUST00000072801.5
ENSMUST00000227672.2 |
Vmn1r82
|
vomeronasal 1 receptor 82 |
| chr4_-_58553553 | 0.10 |
ENSMUST00000107575.9
ENSMUST00000107574.8 ENSMUST00000147354.8 |
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr5_+_92535705 | 0.10 |
ENSMUST00000138687.2
ENSMUST00000124509.2 |
Art3
|
ADP-ribosyltransferase 3 |
| chr3_+_96537484 | 0.09 |
ENSMUST00000200647.2
|
Rbm8a
|
RNA binding motif protein 8a |
| chr5_-_28672091 | 0.09 |
ENSMUST00000002708.5
|
Shh
|
sonic hedgehog |
| chr4_-_49597837 | 0.09 |
ENSMUST00000042750.3
|
Pgap4
|
post-GPI attachment to proteins GalNAc transferase 4 |
| chr7_+_25000836 | 0.09 |
ENSMUST00000080288.5
|
Prr19
|
proline rich 19 |
| chr18_-_78249612 | 0.09 |
ENSMUST00000163367.3
|
Slc14a2
|
solute carrier family 14 (urea transporter), member 2 |
| chr7_-_12418918 | 0.09 |
ENSMUST00000167771.2
ENSMUST00000172743.9 ENSMUST00000233763.2 ENSMUST00000232880.2 |
Vmn2r55
|
vomeronasal 2, receptor 55 |
| chr16_+_36648728 | 0.09 |
ENSMUST00000114819.8
ENSMUST00000023535.4 |
Iqcb1
|
IQ calmodulin-binding motif containing 1 |
| chr7_+_113113292 | 0.09 |
ENSMUST00000122890.2
|
Far1
|
fatty acyl CoA reductase 1 |
| chr3_+_96537235 | 0.09 |
ENSMUST00000048915.11
ENSMUST00000196456.5 ENSMUST00000198027.5 |
Rbm8a
|
RNA binding motif protein 8a |
| chr19_-_13827773 | 0.09 |
ENSMUST00000215350.2
|
Olfr1501
|
olfactory receptor 1501 |
| chr3_-_95158360 | 0.09 |
ENSMUST00000098871.11
ENSMUST00000137250.9 |
Bnipl
|
BCL2/adenovirus E1B 19kD interacting protein like |
| chr2_-_45001141 | 0.09 |
ENSMUST00000201969.4
ENSMUST00000201623.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr7_-_126975169 | 0.09 |
ENSMUST00000205832.2
|
Zfp747
|
zinc finger protein 747 |
| chr19_-_11838430 | 0.09 |
ENSMUST00000214796.2
|
Olfr1418
|
olfactory receptor 1418 |
| chr6_-_34294377 | 0.09 |
ENSMUST00000154655.2
ENSMUST00000102980.11 |
Akr1b3
|
aldo-keto reductase family 1, member B3 (aldose reductase) |
| chr5_-_8417982 | 0.09 |
ENSMUST00000088761.11
ENSMUST00000115386.8 ENSMUST00000050166.14 ENSMUST00000046838.14 ENSMUST00000115388.9 ENSMUST00000088744.12 ENSMUST00000115385.2 |
Adam22
|
a disintegrin and metallopeptidase domain 22 |
| chr9_+_39657545 | 0.08 |
ENSMUST00000213358.2
|
Olfr967
|
olfactory receptor 967 |
| chr19_-_12018681 | 0.08 |
ENSMUST00000214472.2
|
Olfr1423
|
olfactory receptor 1423 |
| chr8_+_94941192 | 0.08 |
ENSMUST00000079961.14
ENSMUST00000212824.2 |
Nup93
|
nucleoporin 93 |
| chr15_-_100322934 | 0.08 |
ENSMUST00000123461.8
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr10_-_23968192 | 0.08 |
ENSMUST00000092654.4
|
Taar8b
|
trace amine-associated receptor 8B |
| chr15_-_100322089 | 0.08 |
ENSMUST00000154331.2
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr5_+_92535532 | 0.08 |
ENSMUST00000145072.8
|
Art3
|
ADP-ribosyltransferase 3 |
| chr14_-_30850881 | 0.08 |
ENSMUST00000203261.3
|
Smim4
|
small integral membrane protein 4 |
| chr4_-_127224591 | 0.08 |
ENSMUST00000046532.4
|
Gjb3
|
gap junction protein, beta 3 |
| chr14_+_99337311 | 0.08 |
ENSMUST00000022650.9
|
Pibf1
|
progesterone immunomodulatory binding factor 1 |
| chr7_-_104542833 | 0.07 |
ENSMUST00000081116.2
|
Olfr666
|
olfactory receptor 666 |
| chr17_-_47813201 | 0.07 |
ENSMUST00000233174.2
ENSMUST00000233121.2 ENSMUST00000067103.4 |
Taf8
|
TATA-box binding protein associated factor 8 |
| chr7_+_113113037 | 0.07 |
ENSMUST00000033018.15
|
Far1
|
fatty acyl CoA reductase 1 |
| chrX_-_58211440 | 0.07 |
ENSMUST00000119306.2
|
Fgf13
|
fibroblast growth factor 13 |
| chr5_+_146885450 | 0.07 |
ENSMUST00000146511.8
ENSMUST00000132102.2 |
Gtf3a
|
general transcription factor III A |
| chr11_-_58353096 | 0.07 |
ENSMUST00000215691.2
|
Olfr30
|
olfactory receptor 30 |
| chr19_+_8595369 | 0.07 |
ENSMUST00000010250.4
|
Slc22a6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr19_-_4861536 | 0.07 |
ENSMUST00000179909.8
ENSMUST00000172000.3 ENSMUST00000180008.2 ENSMUST00000113793.4 ENSMUST00000006625.8 |
Gm21992
Rbm14
|
predicted gene 21992 RNA binding motif protein 14 |
| chr4_+_132495636 | 0.06 |
ENSMUST00000102561.11
|
Rpa2
|
replication protein A2 |
| chr13_+_23966524 | 0.06 |
ENSMUST00000074067.4
|
Trim38
|
tripartite motif-containing 38 |
| chr4_-_43000450 | 0.06 |
ENSMUST00000030164.8
|
Vcp
|
valosin containing protein |
| chr7_+_57069417 | 0.06 |
ENSMUST00000085240.11
|
Gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 3 |
| chr9_-_110453427 | 0.06 |
ENSMUST00000196876.2
ENSMUST00000035069.14 |
Nradd
|
neurotrophin receptor associated death domain |
| chr15_+_89452529 | 0.06 |
ENSMUST00000023295.3
ENSMUST00000230538.2 ENSMUST00000230978.2 |
Acr
|
acrosin prepropeptide |
| chr1_+_74401267 | 0.06 |
ENSMUST00000097697.8
|
Catip
|
ciliogenesis associated TTC17 interacting protein |
| chrX_+_152341587 | 0.06 |
ENSMUST00000112573.8
ENSMUST00000056754.4 |
Cypt3
|
cysteine-rich perinuclear theca 3 |
| chr11_-_69791712 | 0.06 |
ENSMUST00000108621.9
ENSMUST00000100969.9 |
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr11_+_16207705 | 0.06 |
ENSMUST00000109645.9
ENSMUST00000109647.3 |
Vstm2a
|
V-set and transmembrane domain containing 2A |
| chr13_-_21765822 | 0.06 |
ENSMUST00000206526.3
ENSMUST00000213912.2 |
Olfr1364
|
olfactory receptor 1364 |
| chr11_-_69791756 | 0.06 |
ENSMUST00000018714.13
ENSMUST00000128046.2 |
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr11_-_69791774 | 0.05 |
ENSMUST00000102580.10
|
2810408A11Rik
|
RIKEN cDNA 2810408A11 gene |
| chr19_+_12839106 | 0.05 |
ENSMUST00000059675.4
|
Olfr1444
|
olfactory receptor 1444 |
| chr13_+_51799268 | 0.05 |
ENSMUST00000075853.6
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr4_-_43710231 | 0.05 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr16_-_45544960 | 0.05 |
ENSMUST00000096057.5
|
Tagln3
|
transgelin 3 |
| chr5_+_33815466 | 0.05 |
ENSMUST00000074849.13
ENSMUST00000079534.11 ENSMUST00000201633.2 |
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr10_+_39488930 | 0.05 |
ENSMUST00000019987.7
|
Traf3ip2
|
TRAF3 interacting protein 2 |
| chr4_+_116078787 | 0.05 |
ENSMUST00000147292.8
|
Pik3r3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr2_-_57942844 | 0.05 |
ENSMUST00000090940.6
|
Ermn
|
ermin, ERM-like protein |
| chr5_+_57875309 | 0.05 |
ENSMUST00000191837.6
ENSMUST00000068110.10 |
Pcdh7
|
protocadherin 7 |
| chr18_+_42644552 | 0.05 |
ENSMUST00000237602.2
ENSMUST00000236088.2 ENSMUST00000025375.15 |
Tcerg1
|
transcription elongation regulator 1 (CA150) |
| chr16_-_92196954 | 0.04 |
ENSMUST00000023672.10
|
Rcan1
|
regulator of calcineurin 1 |
| chr14_+_65504151 | 0.04 |
ENSMUST00000169656.3
ENSMUST00000226005.2 |
Fbxo16
|
F-box protein 16 |
| chr6_+_63232955 | 0.04 |
ENSMUST00000095852.5
|
Grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr13_+_96524802 | 0.04 |
ENSMUST00000099295.6
|
Poc5
|
POC5 centriolar protein |
| chr5_-_92190859 | 0.04 |
ENSMUST00000069937.11
ENSMUST00000086978.12 |
Cdkl2
|
cyclin-dependent kinase-like 2 (CDC2-related kinase) |
| chr3_-_129518723 | 0.04 |
ENSMUST00000199615.5
ENSMUST00000197079.5 |
Egf
|
epidermal growth factor |
| chr5_+_135197228 | 0.04 |
ENSMUST00000111187.10
ENSMUST00000111188.5 ENSMUST00000202606.3 |
Bcl7b
|
B cell CLL/lymphoma 7B |
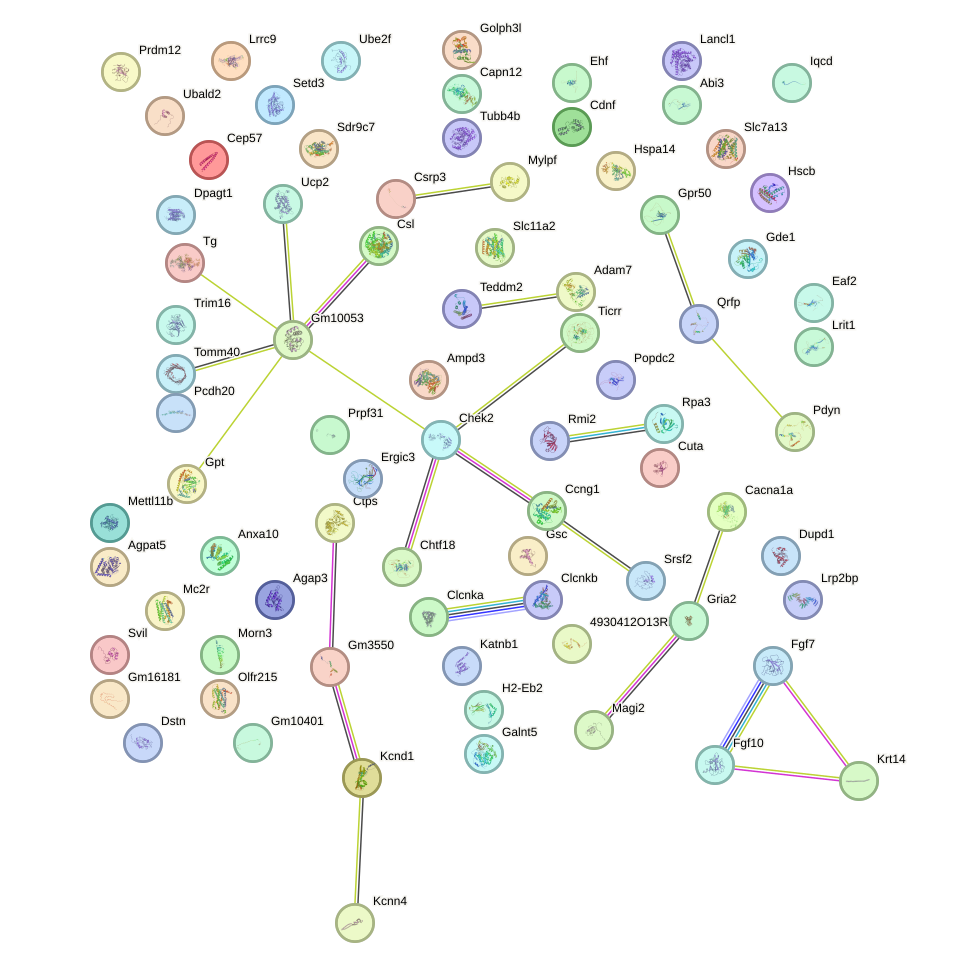
Network of associatons between targets according to the STRING database.
First level regulatory network of Barhl2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 0.6 | GO:1903920 | positive regulation of actin filament severing(GO:1903920) |
| 0.2 | 0.6 | GO:0051329 | interphase(GO:0051325) mitotic interphase(GO:0051329) |
| 0.2 | 1.5 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.1 | 0.4 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.1 | 0.4 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.1 | 0.3 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.1 | 0.4 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.2 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.1 | 0.3 | GO:0015676 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 | 0.2 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.0 | 0.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.3 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.3 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.0 | 0.1 | GO:2000729 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) kidney smooth muscle tissue development(GO:0072194) negative regulation of dopaminergic neuron differentiation(GO:1904339) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.5 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.4 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.2 | GO:0072053 | renal inner medulla development(GO:0072053) |
| 0.0 | 0.1 | GO:0072061 | hexitol metabolic process(GO:0006059) naphthalene metabolic process(GO:0018931) inner medullary collecting duct development(GO:0072061) naphthalene-containing compound metabolic process(GO:0090420) |
| 0.0 | 0.2 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.2 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:2000373 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.0 | 0.1 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.3 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.2 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.0 | 0.1 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.0 | 0.1 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.6 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.2 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.5 | GO:0042761 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.2 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.2 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.0 | 0.1 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.2 | GO:0046036 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.0 | 0.1 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.1 | 0.3 | GO:0002945 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.1 | 0.6 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 0.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.3 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.2 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.1 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.2 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.5 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0004021 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.1 | 1.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.6 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.2 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.1 | 0.4 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 0.4 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.6 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 0.3 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.5 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 1.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.4 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.6 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID ATM PATHWAY | ATM pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.6 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 2.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.5 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.4 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |