Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Cdc5l
Z-value: 0.29
Transcription factors associated with Cdc5l
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cdc5l
|
ENSMUSG00000023932.10 | cell division cycle 5-like (S. pombe) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cdc5l | mm39_v1_chr17_-_45744637_45744671 | -0.23 | 1.9e-01 | Click! |
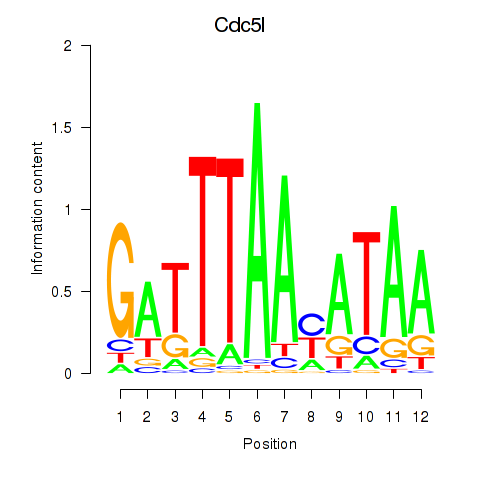
Activity profile of Cdc5l motif
Sorted Z-values of Cdc5l motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_58645189 | 0.92 |
ENSMUST00000102755.4
ENSMUST00000230627.2 ENSMUST00000229923.2 |
Upp2
|
uridine phosphorylase 2 |
| chr7_+_51537645 | 0.56 |
ENSMUST00000208711.2
|
Gas2
|
growth arrest specific 2 |
| chr5_-_136911969 | 0.34 |
ENSMUST00000057497.13
ENSMUST00000111103.2 |
Col26a1
|
collagen, type XXVI, alpha 1 |
| chr8_+_110717062 | 0.31 |
ENSMUST00000001720.14
ENSMUST00000143741.2 |
Tat
|
tyrosine aminotransferase |
| chr3_-_82957104 | 0.30 |
ENSMUST00000048246.5
|
Fgb
|
fibrinogen beta chain |
| chr3_+_137923521 | 0.27 |
ENSMUST00000090171.7
|
Adh7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr10_+_17598961 | 0.24 |
ENSMUST00000038107.9
ENSMUST00000219558.2 ENSMUST00000218370.2 |
Cited2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr14_+_66872699 | 0.24 |
ENSMUST00000159365.8
ENSMUST00000054661.8 ENSMUST00000225182.2 ENSMUST00000159068.2 |
Adra1a
|
adrenergic receptor, alpha 1a |
| chr2_+_125876883 | 0.24 |
ENSMUST00000110442.2
|
Fgf7
|
fibroblast growth factor 7 |
| chr7_-_101517874 | 0.21 |
ENSMUST00000150184.2
|
Folr1
|
folate receptor 1 (adult) |
| chr15_-_13173736 | 0.20 |
ENSMUST00000036439.6
|
Cdh6
|
cadherin 6 |
| chr7_-_101518217 | 0.20 |
ENSMUST00000123321.8
|
Folr1
|
folate receptor 1 (adult) |
| chrX_-_50031587 | 0.19 |
ENSMUST00000060650.7
|
Frmd7
|
FERM domain containing 7 |
| chr3_-_122910662 | 0.17 |
ENSMUST00000051443.8
|
Synpo2
|
synaptopodin 2 |
| chr7_-_104611827 | 0.15 |
ENSMUST00000214216.2
|
Olfr670
|
olfactory receptor 670 |
| chr12_+_59176506 | 0.15 |
ENSMUST00000175912.8
ENSMUST00000176892.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr2_+_125876566 | 0.14 |
ENSMUST00000064794.14
|
Fgf7
|
fibroblast growth factor 7 |
| chr11_+_97732108 | 0.14 |
ENSMUST00000155954.3
ENSMUST00000164364.2 ENSMUST00000170806.2 |
B230217C12Rik
|
RIKEN cDNA B230217C12 gene |
| chr9_-_112016966 | 0.13 |
ENSMUST00000178410.2
ENSMUST00000172380.10 |
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr1_+_143516402 | 0.13 |
ENSMUST00000038252.4
|
B3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chrX_-_93410383 | 0.12 |
ENSMUST00000044989.12
|
Fam90a1b
|
family with sequence similarity 90, member A1B |
| chr12_+_59176543 | 0.12 |
ENSMUST00000069430.15
ENSMUST00000177370.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr7_-_103390529 | 0.12 |
ENSMUST00000051346.5
|
Olfr629
|
olfactory receptor 629 |
| chr11_+_59197746 | 0.11 |
ENSMUST00000000128.10
ENSMUST00000108783.4 |
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr8_+_91681898 | 0.11 |
ENSMUST00000209746.2
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr9_-_112016834 | 0.11 |
ENSMUST00000111872.9
ENSMUST00000164754.9 |
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21 |
| chr5_-_138278223 | 0.11 |
ENSMUST00000014089.9
ENSMUST00000161827.8 |
Gpc2
|
glypican 2 (cerebroglycan) |
| chr14_-_55204023 | 0.10 |
ENSMUST00000124930.8
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr10_+_23814470 | 0.10 |
ENSMUST00000079134.5
|
Taar2
|
trace amine-associated receptor 2 |
| chr10_+_52267702 | 0.10 |
ENSMUST00000067085.7
|
Nepn
|
nephrocan |
| chr5_+_88073483 | 0.09 |
ENSMUST00000113271.3
|
Csn3
|
casein kappa |
| chr5_+_88073438 | 0.09 |
ENSMUST00000001667.13
ENSMUST00000113267.8 |
Csn3
|
casein kappa |
| chr15_-_50746202 | 0.09 |
ENSMUST00000184885.8
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr6_+_6863269 | 0.09 |
ENSMUST00000171311.8
ENSMUST00000160937.9 |
Dlx6
|
distal-less homeobox 6 |
| chr5_+_67125902 | 0.09 |
ENSMUST00000127184.8
|
Limch1
|
LIM and calponin homology domains 1 |
| chr14_-_55204383 | 0.09 |
ENSMUST00000111456.2
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr10_+_12881087 | 0.09 |
ENSMUST00000105139.5
|
Sf3b5
|
splicing factor 3b, subunit 5 |
| chr7_+_5086702 | 0.09 |
ENSMUST00000208634.2
|
Epn1
|
epsin 1 |
| chr1_+_60785517 | 0.09 |
ENSMUST00000027165.3
|
Cd28
|
CD28 antigen |
| chr18_-_22983794 | 0.09 |
ENSMUST00000092015.11
ENSMUST00000069215.13 |
Nol4
|
nucleolar protein 4 |
| chr11_-_78642480 | 0.09 |
ENSMUST00000059468.6
|
Ccnq
|
cyclin Q |
| chr10_-_107747995 | 0.08 |
ENSMUST00000165341.5
|
Otogl
|
otogelin-like |
| chr3_+_93376058 | 0.08 |
ENSMUST00000029516.3
|
Tchhl1
|
trichohyalin-like 1 |
| chr5_+_104447037 | 0.08 |
ENSMUST00000031246.9
|
Ibsp
|
integrin binding sialoprotein |
| chr17_+_47451868 | 0.08 |
ENSMUST00000190080.9
|
Trerf1
|
transcriptional regulating factor 1 |
| chr2_+_106525938 | 0.08 |
ENSMUST00000016530.14
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr5_-_86666408 | 0.08 |
ENSMUST00000140095.2
ENSMUST00000134179.8 |
Tmprss11g
|
transmembrane protease, serine 11g |
| chr14_-_55204054 | 0.07 |
ENSMUST00000226297.2
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr1_+_106866678 | 0.07 |
ENSMUST00000112724.3
|
Serpinb12
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 12 |
| chr5_+_92535705 | 0.07 |
ENSMUST00000138687.2
ENSMUST00000124509.2 |
Art3
|
ADP-ribosyltransferase 3 |
| chr5_+_67125759 | 0.07 |
ENSMUST00000238993.2
ENSMUST00000038188.14 |
Limch1
|
LIM and calponin homology domains 1 |
| chr10_+_20046748 | 0.07 |
ENSMUST00000215924.2
|
Map7
|
microtubule-associated protein 7 |
| chr14_-_55204092 | 0.06 |
ENSMUST00000081857.14
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr4_-_149211145 | 0.06 |
ENSMUST00000030815.3
|
Cort
|
cortistatin |
| chrX_+_135723420 | 0.06 |
ENSMUST00000033800.13
|
Plp1
|
proteolipid protein (myelin) 1 |
| chr9_+_19533374 | 0.05 |
ENSMUST00000213725.2
ENSMUST00000208694.2 |
Zfp317
|
zinc finger protein 317 |
| chr19_+_23881821 | 0.05 |
ENSMUST00000237688.2
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chr12_+_29578354 | 0.05 |
ENSMUST00000218583.2
ENSMUST00000049784.17 |
Myt1l
|
myelin transcription factor 1-like |
| chr4_-_119241024 | 0.05 |
ENSMUST00000127149.8
ENSMUST00000152879.9 ENSMUST00000238673.2 ENSMUST00000238485.2 |
Ccdc30
|
coiled-coil domain containing 30 |
| chr6_+_88701810 | 0.05 |
ENSMUST00000089449.5
|
Mgll
|
monoglyceride lipase |
| chr5_+_18167547 | 0.05 |
ENSMUST00000030561.9
|
Gnat3
|
guanine nucleotide binding protein, alpha transducing 3 |
| chr4_-_119241002 | 0.04 |
ENSMUST00000238721.2
|
Ccdc30
|
coiled-coil domain containing 30 |
| chrX_+_135723531 | 0.04 |
ENSMUST00000113085.2
|
Plp1
|
proteolipid protein (myelin) 1 |
| chr1_+_179938904 | 0.04 |
ENSMUST00000145181.2
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr2_-_62404195 | 0.03 |
ENSMUST00000174234.8
ENSMUST00000000402.16 ENSMUST00000174448.8 ENSMUST00000102732.10 |
Fap
|
fibroblast activation protein |
| chr1_+_17672117 | 0.03 |
ENSMUST00000088476.4
|
Pi15
|
peptidase inhibitor 15 |
| chr7_-_29898236 | 0.02 |
ENSMUST00000001845.13
|
Capns1
|
calpain, small subunit 1 |
| chr2_+_121991181 | 0.02 |
ENSMUST00000036089.8
|
Trim69
|
tripartite motif-containing 69 |
| chr2_+_65451100 | 0.02 |
ENSMUST00000144254.6
ENSMUST00000028377.14 |
Scn2a
|
sodium channel, voltage-gated, type II, alpha |
| chr5_-_31377847 | 0.02 |
ENSMUST00000202294.4
ENSMUST00000031032.11 |
Ppm1g
|
protein phosphatase 1G (formerly 2C), magnesium-dependent, gamma isoform |
| chr5_+_107656810 | 0.01 |
ENSMUST00000160160.6
|
Gm42669
|
predicted gene 42669 |
| chr9_+_19533591 | 0.01 |
ENSMUST00000215372.2
|
Zfp317
|
zinc finger protein 317 |
| chr6_-_14755249 | 0.01 |
ENSMUST00000045096.6
|
Ppp1r3a
|
protein phosphatase 1, regulatory subunit 3A |
| chr4_-_119240885 | 0.01 |
ENSMUST00000238422.2
ENSMUST00000238719.2 ENSMUST00000238723.2 ENSMUST00000044781.9 ENSMUST00000084307.5 ENSMUST00000148236.9 ENSMUST00000127474.3 ENSMUST00000238704.2 |
Ccdc30
|
coiled-coil domain containing 30 |
| chr5_+_20433169 | 0.01 |
ENSMUST00000197553.5
ENSMUST00000208219.2 |
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Cdc5l
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.1 | 0.3 | GO:0007522 | visceral muscle development(GO:0007522) |
| 0.1 | 0.2 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.1 | 0.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.3 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.1 | 0.2 | GO:0035802 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) |
| 0.0 | 0.4 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.0 | 0.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.2 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 0.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.1 | 0.3 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.3 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.4 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.3 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.4 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.1 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 0.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.4 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |