Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
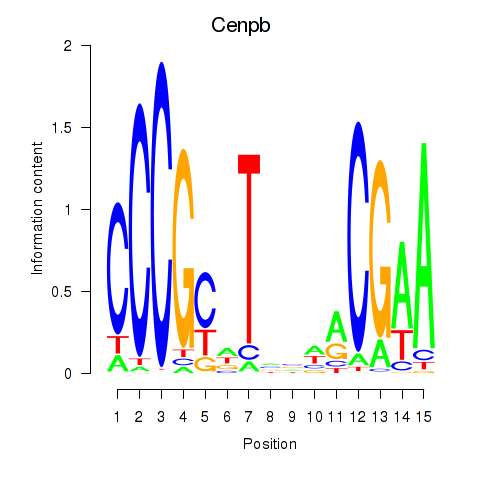
Results for Cenpb
Z-value: 0.73
Transcription factors associated with Cenpb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cenpb
|
ENSMUSG00000068267.6 | centromere protein B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cenpb | mm39_v1_chr2_-_131021905_131022005 | -0.09 | 5.9e-01 | Click! |
Activity profile of Cenpb motif
Sorted Z-values of Cenpb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_41012435 | 4.36 |
ENSMUST00000031931.6
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr3_-_113263974 | 3.19 |
ENSMUST00000098667.5
|
Amy2a2
|
amylase 2a2 |
| chr1_+_171041539 | 2.01 |
ENSMUST00000005820.11
ENSMUST00000075469.12 ENSMUST00000155126.8 |
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr1_+_171041583 | 1.94 |
ENSMUST00000111328.8
|
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr15_-_100579813 | 1.86 |
ENSMUST00000230572.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr14_-_44057096 | 1.76 |
ENSMUST00000100691.4
|
Ear1
|
eosinophil-associated, ribonuclease A family, member 1 |
| chr9_+_48406706 | 1.67 |
ENSMUST00000048824.9
|
Gm5617
|
predicted gene 5617 |
| chr5_-_140687995 | 1.60 |
ENSMUST00000135028.5
ENSMUST00000077890.12 ENSMUST00000041783.14 ENSMUST00000142081.6 |
Iqce
|
IQ motif containing E |
| chr14_-_44112974 | 1.22 |
ENSMUST00000179200.2
|
Ear1
|
eosinophil-associated, ribonuclease A family, member 1 |
| chr7_+_125151432 | 1.15 |
ENSMUST00000206217.2
ENSMUST00000205985.2 |
Il4ra
|
interleukin 4 receptor, alpha |
| chr5_-_124387812 | 1.10 |
ENSMUST00000162812.8
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr6_-_39534765 | 1.03 |
ENSMUST00000036877.10
ENSMUST00000154149.2 |
Dennd2a
|
DENN/MADD domain containing 2A |
| chr11_+_100973391 | 1.01 |
ENSMUST00000001806.10
ENSMUST00000107308.4 |
Coasy
|
Coenzyme A synthase |
| chr11_+_70104929 | 0.98 |
ENSMUST00000094055.10
ENSMUST00000126296.8 ENSMUST00000136328.2 ENSMUST00000153993.3 |
Slc16a11
|
solute carrier family 16 (monocarboxylic acid transporters), member 11 |
| chr7_+_125151292 | 0.95 |
ENSMUST00000033004.8
|
Il4ra
|
interleukin 4 receptor, alpha |
| chr6_+_48531710 | 0.90 |
ENSMUST00000114545.8
ENSMUST00000153222.2 ENSMUST00000204071.2 ENSMUST00000101436.3 ENSMUST00000203627.2 |
Lrrc61
|
leucine rich repeat containing 61 |
| chr7_-_19363280 | 0.90 |
ENSMUST00000094762.10
ENSMUST00000049912.15 ENSMUST00000098754.5 |
Relb
|
avian reticuloendotheliosis viral (v-rel) oncogene related B |
| chr17_+_34043536 | 0.89 |
ENSMUST00000048249.8
|
Ndufa7
|
NADH:ubiquinone oxidoreductase subunit A7 |
| chr10_-_95252712 | 0.89 |
ENSMUST00000020215.16
|
Socs2
|
suppressor of cytokine signaling 2 |
| chr2_-_27317004 | 0.88 |
ENSMUST00000056176.8
|
Vav2
|
vav 2 oncogene |
| chr10_+_75242745 | 0.83 |
ENSMUST00000039925.8
|
Upb1
|
ureidopropionase, beta |
| chr4_-_149858694 | 0.81 |
ENSMUST00000105686.3
|
Slc25a33
|
solute carrier family 25, member 33 |
| chr5_-_38659449 | 0.76 |
ENSMUST00000005238.13
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr6_-_94677118 | 0.75 |
ENSMUST00000101126.3
ENSMUST00000032105.11 |
Lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr17_+_30223707 | 0.75 |
ENSMUST00000226208.3
|
Zfand3
|
zinc finger, AN1-type domain 3 |
| chr8_+_84728123 | 0.71 |
ENSMUST00000060357.15
ENSMUST00000239176.2 |
1700067K01Rik
|
RIKEN cDNA 1700067K01 gene |
| chr7_-_140590605 | 0.71 |
ENSMUST00000026565.7
|
Ifitm3
|
interferon induced transmembrane protein 3 |
| chr10_-_81262948 | 0.70 |
ENSMUST00000078185.14
ENSMUST00000020461.15 ENSMUST00000105321.10 |
Nfic
|
nuclear factor I/C |
| chr17_+_74111823 | 0.70 |
ENSMUST00000024860.9
|
Ehd3
|
EH-domain containing 3 |
| chr5_-_100563974 | 0.69 |
ENSMUST00000182886.8
ENSMUST00000094578.11 |
Sec31a
|
Sec31 homolog A (S. cerevisiae) |
| chr2_+_168072519 | 0.68 |
ENSMUST00000099071.5
|
Mocs3
|
molybdenum cofactor synthesis 3 |
| chr11_-_120348762 | 0.68 |
ENSMUST00000137632.2
ENSMUST00000044007.3 |
Oxld1
|
oxidoreductase like domain containing 1 |
| chr17_-_35827676 | 0.65 |
ENSMUST00000160885.2
ENSMUST00000159009.2 ENSMUST00000161012.8 |
Tcf19
|
transcription factor 19 |
| chr1_-_155688551 | 0.65 |
ENSMUST00000194632.2
ENSMUST00000111764.8 |
Qsox1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr5_+_100993996 | 0.63 |
ENSMUST00000112887.8
ENSMUST00000031255.15 |
Gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr5_-_38659422 | 0.63 |
ENSMUST00000147664.8
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr9_+_107173907 | 0.60 |
ENSMUST00000168260.2
|
Cish
|
cytokine inducible SH2-containing protein |
| chr9_+_95441652 | 0.58 |
ENSMUST00000079597.7
|
Paqr9
|
progestin and adipoQ receptor family member IX |
| chr8_+_92040215 | 0.58 |
ENSMUST00000166548.9
|
Fto
|
fat mass and obesity associated |
| chr8_+_92040188 | 0.58 |
ENSMUST00000136802.8
|
Fto
|
fat mass and obesity associated |
| chr9_+_44966464 | 0.57 |
ENSMUST00000114664.8
|
Mpzl3
|
myelin protein zero-like 3 |
| chr9_-_122123475 | 0.55 |
ENSMUST00000042546.4
|
Ano10
|
anoctamin 10 |
| chr14_-_61283911 | 0.55 |
ENSMUST00000111234.10
ENSMUST00000224371.2 |
Tnfrsf19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr6_-_4747157 | 0.55 |
ENSMUST00000126151.8
ENSMUST00000115577.9 ENSMUST00000101677.9 ENSMUST00000115579.8 ENSMUST00000004750.15 |
Sgce
|
sarcoglycan, epsilon |
| chr8_+_70755168 | 0.55 |
ENSMUST00000066469.14
|
Cope
|
coatomer protein complex, subunit epsilon |
| chr8_-_84874468 | 0.54 |
ENSMUST00000117424.9
ENSMUST00000040383.9 |
Cc2d1a
|
coiled-coil and C2 domain containing 1A |
| chr5_+_100994230 | 0.54 |
ENSMUST00000092990.4
ENSMUST00000145612.2 |
Gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr2_-_39116044 | 0.53 |
ENSMUST00000204368.2
|
Ppp6c
|
protein phosphatase 6, catalytic subunit |
| chr6_-_88852017 | 0.53 |
ENSMUST00000145944.3
|
Podxl2
|
podocalyxin-like 2 |
| chr2_-_122199643 | 0.53 |
ENSMUST00000125826.8
|
Shf
|
Src homology 2 domain containing F |
| chr11_-_120238917 | 0.52 |
ENSMUST00000106215.11
|
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr9_-_122123427 | 0.52 |
ENSMUST00000216670.2
|
Ano10
|
anoctamin 10 |
| chr2_-_122199604 | 0.52 |
ENSMUST00000151130.8
|
Shf
|
Src homology 2 domain containing F |
| chr15_-_89033761 | 0.51 |
ENSMUST00000088823.5
|
Mapk11
|
mitogen-activated protein kinase 11 |
| chr17_+_21165573 | 0.51 |
ENSMUST00000007708.14
|
Ppp2r1a
|
protein phosphatase 2, regulatory subunit A, alpha |
| chr17_-_26011241 | 0.50 |
ENSMUST00000237093.2
|
Antkmt
|
adenine nucleotide translocase lysine methyltransferase |
| chr5_+_29774437 | 0.50 |
ENSMUST00000199032.2
|
Ube3c
|
ubiquitin protein ligase E3C |
| chr6_-_4747066 | 0.50 |
ENSMUST00000090686.11
ENSMUST00000133306.8 |
Sgce
|
sarcoglycan, epsilon |
| chr8_+_127790772 | 0.50 |
ENSMUST00000079777.12
ENSMUST00000160272.8 ENSMUST00000162907.8 ENSMUST00000162536.8 ENSMUST00000026921.13 ENSMUST00000162665.8 ENSMUST00000162602.8 ENSMUST00000160581.8 ENSMUST00000161355.8 ENSMUST00000162531.8 ENSMUST00000160766.8 ENSMUST00000159537.8 |
Pard3
|
par-3 family cell polarity regulator |
| chr13_+_91889626 | 0.49 |
ENSMUST00000022120.5
|
Acot12
|
acyl-CoA thioesterase 12 |
| chr16_+_17577464 | 0.48 |
ENSMUST00000129199.8
|
Klhl22
|
kelch-like 22 |
| chr6_+_115521625 | 0.48 |
ENSMUST00000130425.8
ENSMUST00000040234.9 |
Tsen2
|
tRNA splicing endonuclease subunit 2 |
| chr1_-_183766195 | 0.47 |
ENSMUST00000050306.8
|
1700056E22Rik
|
RIKEN cDNA 1700056E22 gene |
| chr11_+_115656246 | 0.45 |
ENSMUST00000093912.11
ENSMUST00000136720.8 ENSMUST00000103034.10 ENSMUST00000141871.8 |
Tmem94
|
transmembrane protein 94 |
| chr8_+_71358576 | 0.45 |
ENSMUST00000019405.4
ENSMUST00000212511.2 |
Map1s
|
microtubule-associated protein 1S |
| chr17_-_26011357 | 0.45 |
ENSMUST00000236683.2
|
Antkmt
|
adenine nucleotide translocase lysine methyltransferase |
| chr14_+_66534539 | 0.45 |
ENSMUST00000121006.2
|
Trim35
|
tripartite motif-containing 35 |
| chr7_-_44541787 | 0.45 |
ENSMUST00000208551.2
ENSMUST00000208253.2 ENSMUST00000207654.2 ENSMUST00000207278.2 |
Med25
|
mediator complex subunit 25 |
| chr13_+_55300453 | 0.45 |
ENSMUST00000005452.6
|
Fgfr4
|
fibroblast growth factor receptor 4 |
| chr2_+_127178072 | 0.45 |
ENSMUST00000028846.7
|
Dusp2
|
dual specificity phosphatase 2 |
| chr11_-_116089595 | 0.45 |
ENSMUST00000072948.11
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr11_+_101137231 | 0.44 |
ENSMUST00000122006.8
ENSMUST00000151830.2 |
Ramp2
|
receptor (calcitonin) activity modifying protein 2 |
| chr16_-_94327689 | 0.44 |
ENSMUST00000023615.7
|
Vps26c
|
VPS26 endosomal protein sorting factor C |
| chr1_-_184731672 | 0.44 |
ENSMUST00000192657.2
ENSMUST00000027929.10 |
Mark1
|
MAP/microtubule affinity regulating kinase 1 |
| chr4_+_135691030 | 0.44 |
ENSMUST00000102541.10
ENSMUST00000149636.2 ENSMUST00000143304.2 |
Gale
|
galactose-4-epimerase, UDP |
| chr7_+_28937898 | 0.43 |
ENSMUST00000138128.3
ENSMUST00000142519.3 |
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr15_-_81900335 | 0.43 |
ENSMUST00000152227.8
|
Desi1
|
desumoylating isopeptidase 1 |
| chr7_+_28937859 | 0.43 |
ENSMUST00000108237.2
|
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr9_-_71678814 | 0.43 |
ENSMUST00000122065.2
ENSMUST00000121322.8 ENSMUST00000072899.9 |
Cgnl1
|
cingulin-like 1 |
| chr8_+_3543131 | 0.42 |
ENSMUST00000061508.8
ENSMUST00000207318.2 |
Zfp358
|
zinc finger protein 358 |
| chr8_+_92040153 | 0.41 |
ENSMUST00000069718.15
ENSMUST00000125471.8 ENSMUST00000128081.8 |
Fto
|
fat mass and obesity associated |
| chr17_-_36290571 | 0.41 |
ENSMUST00000173724.2
ENSMUST00000172900.8 ENSMUST00000174849.8 |
Prr3
|
proline-rich polypeptide 3 |
| chr10_-_95253042 | 0.41 |
ENSMUST00000135822.8
|
Socs2
|
suppressor of cytokine signaling 2 |
| chr17_+_24645615 | 0.41 |
ENSMUST00000234304.2
ENSMUST00000024946.7 ENSMUST00000234150.2 |
Eci1
|
enoyl-Coenzyme A delta isomerase 1 |
| chr10_+_80591030 | 0.41 |
ENSMUST00000105336.9
|
Dot1l
|
DOT1-like, histone H3 methyltransferase (S. cerevisiae) |
| chr2_+_157120946 | 0.38 |
ENSMUST00000116380.9
ENSMUST00000029171.6 |
Rpn2
|
ribophorin II |
| chr16_-_28748410 | 0.38 |
ENSMUST00000100023.3
|
Mb21d2
|
Mab-21 domain containing 2 |
| chr6_-_31540913 | 0.38 |
ENSMUST00000026698.8
|
Podxl
|
podocalyxin-like |
| chr11_+_101137786 | 0.38 |
ENSMUST00000107282.4
|
Ramp2
|
receptor (calcitonin) activity modifying protein 2 |
| chr16_+_91855158 | 0.37 |
ENSMUST00000047429.9
ENSMUST00000232677.2 ENSMUST00000113975.3 |
Mrps6
Gm49711
Slc5a3
|
mitochondrial ribosomal protein S6 predicted gene, 49711 solute carrier family 5 (inositol transporters), member 3 |
| chr1_+_180769890 | 0.37 |
ENSMUST00000161847.8
|
Tmem63a
|
transmembrane protein 63a |
| chr17_+_36290743 | 0.37 |
ENSMUST00000087200.4
|
Gnl1
|
guanine nucleotide binding protein-like 1 |
| chr6_+_48715971 | 0.36 |
ENSMUST00000054368.7
ENSMUST00000140054.3 ENSMUST00000204168.2 ENSMUST00000204408.2 |
Gimap1
Gm28053
|
GTPase, IMAP family member 1 predicted gene, 28053 |
| chr8_+_84874654 | 0.36 |
ENSMUST00000143833.8
ENSMUST00000118856.8 |
Brme1
|
break repair meiotic recombinase recruitment factor 1 |
| chr7_+_141041637 | 0.35 |
ENSMUST00000167491.8
ENSMUST00000165194.2 |
Cracr2b
|
calcium release activated channel regulator 2B |
| chr9_+_57468217 | 0.34 |
ENSMUST00000045791.11
ENSMUST00000216986.2 |
Scamp2
|
secretory carrier membrane protein 2 |
| chrX_+_20483742 | 0.34 |
ENSMUST00000115375.8
ENSMUST00000115374.8 ENSMUST00000084383.10 |
Rbm10
|
RNA binding motif protein 10 |
| chr3_-_96812610 | 0.34 |
ENSMUST00000029738.14
|
Gpr89
|
G protein-coupled receptor 89 |
| chr8_-_71112295 | 0.34 |
ENSMUST00000211715.2
ENSMUST00000210307.2 ENSMUST00000209768.2 ENSMUST00000070173.9 |
Pgpep1
|
pyroglutamyl-peptidase I |
| chr4_-_108075119 | 0.34 |
ENSMUST00000223127.2
ENSMUST00000043793.7 ENSMUST00000106690.9 |
Zyg11a
|
zyg-11 family member A, cell cycle regulator |
| chr10_-_81121596 | 0.33 |
ENSMUST00000218484.2
|
Tjp3
|
tight junction protein 3 |
| chr7_+_24920840 | 0.33 |
ENSMUST00000055604.6
|
Zfp526
|
zinc finger protein 526 |
| chr12_-_45120895 | 0.33 |
ENSMUST00000120531.8
ENSMUST00000143376.8 |
Stxbp6
|
syntaxin binding protein 6 (amisyn) |
| chr14_+_66534478 | 0.32 |
ENSMUST00000022623.13
|
Trim35
|
tripartite motif-containing 35 |
| chr3_+_123061094 | 0.31 |
ENSMUST00000047923.12
ENSMUST00000200333.2 |
Sec24d
|
Sec24 related gene family, member D (S. cerevisiae) |
| chr1_+_180770064 | 0.31 |
ENSMUST00000159436.8
|
Tmem63a
|
transmembrane protein 63a |
| chr2_+_180961507 | 0.31 |
ENSMUST00000098971.11
ENSMUST00000054622.15 ENSMUST00000108814.8 ENSMUST00000048608.16 ENSMUST00000108815.8 |
Rtel1
|
regulator of telomere elongation helicase 1 |
| chr11_-_100036792 | 0.31 |
ENSMUST00000007317.8
|
Krt19
|
keratin 19 |
| chr11_+_58868919 | 0.31 |
ENSMUST00000108809.8
ENSMUST00000108810.10 ENSMUST00000093061.7 |
Trim11
|
tripartite motif-containing 11 |
| chr17_-_24005020 | 0.30 |
ENSMUST00000024704.10
|
Flywch2
|
FLYWCH family member 2 |
| chr15_+_78312851 | 0.30 |
ENSMUST00000159771.8
|
Kctd17
|
potassium channel tetramerisation domain containing 17 |
| chr7_+_28937746 | 0.29 |
ENSMUST00000108238.8
ENSMUST00000032809.10 |
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr11_+_87989972 | 0.29 |
ENSMUST00000018522.13
|
Cuedc1
|
CUE domain containing 1 |
| chr8_+_84874881 | 0.28 |
ENSMUST00000093375.5
|
Brme1
|
break repair meiotic recombinase recruitment factor 1 |
| chr5_-_35732390 | 0.28 |
ENSMUST00000030980.12
|
Trmt44
|
tRNA methyltransferase 44 |
| chr17_+_27000034 | 0.27 |
ENSMUST00000015725.16
ENSMUST00000135824.8 ENSMUST00000137989.2 |
Bnip1
|
BCL2/adenovirus E1B interacting protein 1 |
| chr10_+_25235696 | 0.27 |
ENSMUST00000053748.16
|
Epb41l2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr3_+_89738654 | 0.27 |
ENSMUST00000050401.6
|
She
|
src homology 2 domain-containing transforming protein E |
| chr16_-_97564910 | 0.27 |
ENSMUST00000019386.10
|
Ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chrX_+_93768175 | 0.26 |
ENSMUST00000101388.4
|
Zxdb
|
zinc finger, X-linked, duplicated B |
| chrX_-_100312629 | 0.26 |
ENSMUST00000117736.2
|
Gm20489
|
predicted gene 20489 |
| chr11_+_105956867 | 0.26 |
ENSMUST00000002048.8
|
Taco1
|
translational activator of mitochondrially encoded cytochrome c oxidase I |
| chr2_-_168072295 | 0.26 |
ENSMUST00000154111.8
|
Dpm1
|
dolichol-phosphate (beta-D) mannosyltransferase 1 |
| chr16_-_5021843 | 0.26 |
ENSMUST00000147567.2
ENSMUST00000023911.11 |
Nagpa
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase |
| chr10_+_25235748 | 0.25 |
ENSMUST00000218903.2
|
Epb41l2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr8_+_127790626 | 0.25 |
ENSMUST00000162309.8
|
Pard3
|
par-3 family cell polarity regulator |
| chr2_+_180961599 | 0.24 |
ENSMUST00000153112.2
|
Rtel1
|
regulator of telomere elongation helicase 1 |
| chr7_+_141040988 | 0.23 |
ENSMUST00000053670.12
|
Cracr2b
|
calcium release activated channel regulator 2B |
| chrX_-_142610371 | 0.23 |
ENSMUST00000087316.6
|
Capn6
|
calpain 6 |
| chr17_+_23898633 | 0.23 |
ENSMUST00000233364.2
|
Cldn6
|
claudin 6 |
| chr19_-_5168251 | 0.23 |
ENSMUST00000113728.8
ENSMUST00000113727.8 ENSMUST00000025798.13 |
Klc2
|
kinesin light chain 2 |
| chr17_+_80597632 | 0.22 |
ENSMUST00000227729.2
ENSMUST00000061703.10 |
Morn2
|
MORN repeat containing 2 |
| chr11_+_97594220 | 0.22 |
ENSMUST00000103147.5
|
Psmb3
|
proteasome (prosome, macropain) subunit, beta type 3 |
| chr11_-_60307847 | 0.22 |
ENSMUST00000108721.9
ENSMUST00000239016.2 |
Atpaf2
|
ATP synthase mitochondrial F1 complex assembly factor 2 |
| chr11_+_101339233 | 0.22 |
ENSMUST00000010502.13
|
Ifi35
|
interferon-induced protein 35 |
| chr2_-_39116284 | 0.21 |
ENSMUST00000204701.3
|
Ppp6c
|
protein phosphatase 6, catalytic subunit |
| chr11_-_110142565 | 0.21 |
ENSMUST00000044003.14
|
Abca6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr16_+_17577493 | 0.21 |
ENSMUST00000165790.9
|
Klhl22
|
kelch-like 22 |
| chr15_-_83033471 | 0.21 |
ENSMUST00000129372.2
|
Poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr8_-_72763462 | 0.21 |
ENSMUST00000003574.5
|
Cyp4f18
|
cytochrome P450, family 4, subfamily f, polypeptide 18 |
| chr6_+_58808733 | 0.21 |
ENSMUST00000126292.8
ENSMUST00000031823.12 |
Herc3
|
hect domain and RLD 3 |
| chr7_-_80037688 | 0.20 |
ENSMUST00000080932.8
|
Fes
|
feline sarcoma oncogene |
| chr9_+_55116474 | 0.20 |
ENSMUST00000146201.8
|
Fbxo22
|
F-box protein 22 |
| chr14_+_30973407 | 0.20 |
ENSMUST00000022458.11
|
Bap1
|
Brca1 associated protein 1 |
| chr7_-_5017642 | 0.19 |
ENSMUST00000207412.2
ENSMUST00000077385.15 ENSMUST00000165320.3 |
Fiz1
|
Flt3 interacting zinc finger protein 1 |
| chr7_+_29991101 | 0.19 |
ENSMUST00000150892.2
ENSMUST00000126216.2 ENSMUST00000014065.16 |
Clip3
|
CAP-GLY domain containing linker protein 3 |
| chr11_+_120348919 | 0.19 |
ENSMUST00000058370.14
ENSMUST00000175970.8 ENSMUST00000176120.2 |
Ccdc137
|
coiled-coil domain containing 137 |
| chr8_-_70755132 | 0.19 |
ENSMUST00000008004.10
|
Ddx49
|
DEAD box helicase 49 |
| chr9_+_55116209 | 0.18 |
ENSMUST00000034859.15
|
Fbxo22
|
F-box protein 22 |
| chrX_+_57075981 | 0.18 |
ENSMUST00000088627.11
|
Zic3
|
zinc finger protein of the cerebellum 3 |
| chr2_-_168072493 | 0.18 |
ENSMUST00000109193.8
|
Dpm1
|
dolichol-phosphate (beta-D) mannosyltransferase 1 |
| chr15_-_75793312 | 0.17 |
ENSMUST00000053918.9
|
Pycrl
|
pyrroline-5-carboxylate reductase-like |
| chr10_+_128247598 | 0.17 |
ENSMUST00000096386.13
|
Rnf41
|
ring finger protein 41 |
| chr7_-_27373939 | 0.16 |
ENSMUST00000138243.2
|
Map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr8_+_27468175 | 0.16 |
ENSMUST00000209411.2
|
Zfp703
|
zinc finger protein 703 |
| chr11_+_79551358 | 0.16 |
ENSMUST00000155381.2
|
Rab11fip4
|
RAB11 family interacting protein 4 (class II) |
| chr7_-_80037622 | 0.16 |
ENSMUST00000206698.2
|
Fes
|
feline sarcoma oncogene |
| chr7_+_5017414 | 0.16 |
ENSMUST00000207901.2
|
Zfp524
|
zinc finger protein 524 |
| chr1_-_125363159 | 0.16 |
ENSMUST00000191004.7
|
Actr3
|
ARP3 actin-related protein 3 |
| chr7_-_30428930 | 0.16 |
ENSMUST00000207296.2
ENSMUST00000006478.10 |
Tmem147
|
transmembrane protein 147 |
| chr11_-_113641980 | 0.15 |
ENSMUST00000153453.2
|
Cdc42ep4
|
CDC42 effector protein (Rho GTPase binding) 4 |
| chr16_-_18630722 | 0.15 |
ENSMUST00000000028.14
ENSMUST00000115585.2 |
Cdc45
|
cell division cycle 45 |
| chr11_-_102992434 | 0.15 |
ENSMUST00000103077.2
|
Plcd3
|
phospholipase C, delta 3 |
| chr9_+_108225026 | 0.15 |
ENSMUST00000035237.12
ENSMUST00000194959.6 |
Usp4
|
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr11_-_51647204 | 0.15 |
ENSMUST00000109092.8
ENSMUST00000064297.5 |
Sec24a
|
Sec24 related gene family, member A (S. cerevisiae) |
| chr11_-_113642135 | 0.15 |
ENSMUST00000106616.2
|
Cdc42ep4
|
CDC42 effector protein (Rho GTPase binding) 4 |
| chr15_-_78056618 | 0.15 |
ENSMUST00000229476.2
|
Ift27
|
intraflagellar transport 27 |
| chr17_+_43878989 | 0.15 |
ENSMUST00000167214.8
ENSMUST00000024706.12 |
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr17_-_37262495 | 0.15 |
ENSMUST00000040402.14
ENSMUST00000174711.8 |
Ppp1r11
|
protein phosphatase 1, regulatory inhibitor subunit 11 |
| chr5_-_106606032 | 0.15 |
ENSMUST00000086795.8
|
Barhl2
|
BarH like homeobox 2 |
| chrX_-_100266032 | 0.14 |
ENSMUST00000120389.8
ENSMUST00000156473.8 ENSMUST00000077876.4 |
Snx12
|
sorting nexin 12 |
| chr16_+_51851948 | 0.13 |
ENSMUST00000226593.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr2_-_26242079 | 0.13 |
ENSMUST00000028295.9
|
Dnlz
|
DNL-type zinc finger |
| chr16_+_51851917 | 0.13 |
ENSMUST00000227062.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr17_-_36290129 | 0.13 |
ENSMUST00000165613.9
ENSMUST00000173872.8 |
Prr3
|
proline-rich polypeptide 3 |
| chr11_+_72580823 | 0.12 |
ENSMUST00000155998.2
|
Ankfy1
|
ankyrin repeat and FYVE domain containing 1 |
| chr10_+_34173426 | 0.12 |
ENSMUST00000047935.8
|
Tspyl4
|
TSPY-like 4 |
| chr1_+_75498162 | 0.12 |
ENSMUST00000027414.16
ENSMUST00000113553.2 |
Stk11ip
|
serine/threonine kinase 11 interacting protein |
| chr17_-_34174631 | 0.12 |
ENSMUST00000174609.9
ENSMUST00000008812.9 |
Rps18
|
ribosomal protein S18 |
| chr11_+_114618209 | 0.12 |
ENSMUST00000069325.14
|
Dnai2
|
dynein axonemal intermediate chain 2 |
| chr10_+_128247492 | 0.12 |
ENSMUST00000171342.3
|
Rnf41
|
ring finger protein 41 |
| chr11_+_120375439 | 0.11 |
ENSMUST00000043627.8
|
Mrpl12
|
mitochondrial ribosomal protein L12 |
| chr16_+_80997580 | 0.11 |
ENSMUST00000037785.14
ENSMUST00000067602.5 |
Ncam2
|
neural cell adhesion molecule 2 |
| chr2_-_29983618 | 0.11 |
ENSMUST00000081838.7
ENSMUST00000102865.11 |
Zdhhc12
|
zinc finger, DHHC domain containing 12 |
| chr11_-_62172164 | 0.11 |
ENSMUST00000072916.5
|
Zswim7
|
zinc finger SWIM-type containing 7 |
| chr1_-_125363207 | 0.11 |
ENSMUST00000185280.2
|
Actr3
|
ARP3 actin-related protein 3 |
| chr7_-_30428746 | 0.11 |
ENSMUST00000209065.2
ENSMUST00000208169.2 |
Tmem147
|
transmembrane protein 147 |
| chr17_+_26028059 | 0.11 |
ENSMUST00000045692.9
|
Fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr7_-_80037153 | 0.11 |
ENSMUST00000206728.2
|
Fes
|
feline sarcoma oncogene |
| chr4_+_43578922 | 0.11 |
ENSMUST00000030190.9
|
Rgp1
|
RAB6A GEF compex partner 1 |
| chr17_-_56933872 | 0.11 |
ENSMUST00000047226.10
|
Lonp1
|
lon peptidase 1, mitochondrial |
| chr3_-_116388334 | 0.10 |
ENSMUST00000197190.5
ENSMUST00000198454.2 |
Trmt13
|
tRNA methyltransferase 13 |
| chr9_+_57496725 | 0.10 |
ENSMUST00000053230.7
|
Ulk3
|
unc-51-like kinase 3 |
| chr4_-_132611820 | 0.10 |
ENSMUST00000030698.5
|
Stx12
|
syntaxin 12 |
| chr4_-_133066594 | 0.10 |
ENSMUST00000043305.14
|
Wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr5_+_38196568 | 0.10 |
ENSMUST00000031008.13
ENSMUST00000042146.15 ENSMUST00000202412.4 ENSMUST00000154929.5 |
Stx18
|
syntaxin 18 |
| chr5_-_138169253 | 0.10 |
ENSMUST00000139983.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr17_+_6156738 | 0.09 |
ENSMUST00000142030.8
|
Tulp4
|
tubby like protein 4 |
| chr8_+_13338174 | 0.09 |
ENSMUST00000045229.7
|
Tmco3
|
transmembrane and coiled-coil domains 3 |
| chr4_-_123033721 | 0.09 |
ENSMUST00000030404.5
|
Ppie
|
peptidylprolyl isomerase E (cyclophilin E) |
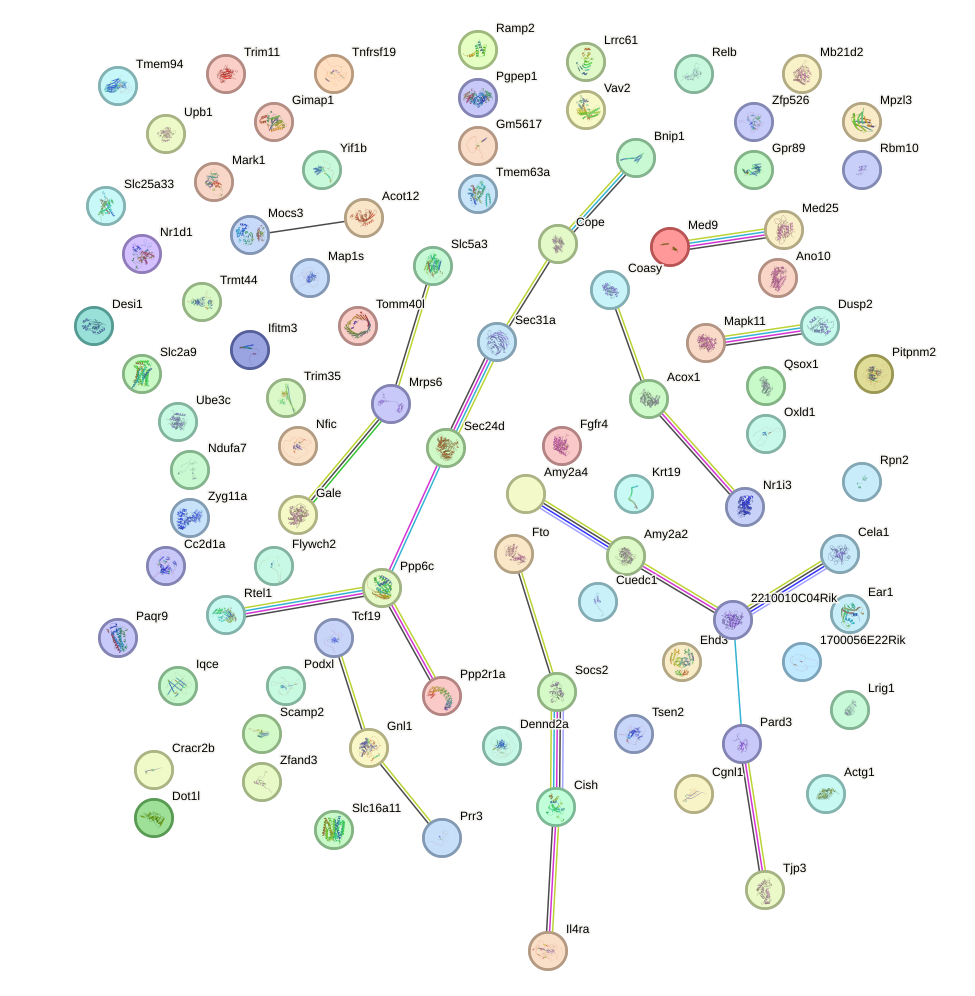
Network of associatons between targets according to the STRING database.
First level regulatory network of Cenpb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.4 | 1.6 | GO:0042245 | RNA repair(GO:0042245) |
| 0.3 | 1.9 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.2 | 0.7 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.2 | 0.5 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.2 | 0.7 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.2 | 0.8 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.2 | 0.8 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.1 | 0.4 | GO:2001178 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.1 | 0.4 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.1 | 0.4 | GO:0061623 | glycolytic process from galactose(GO:0061623) |
| 0.1 | 0.6 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) negative regulation of t-circle formation(GO:1904430) |
| 0.1 | 0.5 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.7 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.8 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 0.4 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.1 | 0.8 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.1 | 0.2 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.1 | 0.4 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.2 | GO:0009233 | menaquinone metabolic process(GO:0009233) phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 | 0.5 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 1.4 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.4 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 0.2 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.1 | 1.0 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.3 | GO:1904171 | negative regulation of bleb assembly(GO:1904171) |
| 0.1 | 1.0 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.2 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.1 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 | 1.1 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.7 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.2 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 1.3 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.5 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.9 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.4 | GO:0015791 | polyol transport(GO:0015791) |
| 0.0 | 0.1 | GO:0097494 | regulation of vesicle size(GO:0097494) |
| 0.0 | 0.2 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.0 | 0.4 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.6 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.3 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.3 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:0032618 | interleukin-15 production(GO:0032618) |
| 0.0 | 0.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.9 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 0.1 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.0 | 0.2 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.6 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.1 | GO:0072299 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.0 | 0.4 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.4 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.2 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.3 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.1 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 0.0 | 0.5 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.6 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.2 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.8 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.5 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 0.0 | 1.2 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.0 | GO:0000239 | pachytene(GO:0000239) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 3.2 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.0 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.0 | 0.1 | GO:0046707 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.0 | 0.9 | GO:0045214 | sarcomere organization(GO:0045214) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.1 | 0.8 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.1 | 1.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.4 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 0.7 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.7 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.5 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.9 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 1.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.2 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0034066 | RIC1-RGP1 guanyl-nucleotide exchange factor complex(GO:0034066) |
| 0.0 | 0.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.4 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.6 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:1990931 | oxidative DNA demethylase activity(GO:0035516) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.5 | 3.2 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.3 | 1.0 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.3 | 4.0 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.2 | 1.3 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 1.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.8 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.1 | 0.6 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.7 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 0.4 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 0.5 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 1.4 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.4 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.1 | 0.5 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 0.3 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 0.2 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 0.1 | 0.4 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.4 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.5 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.8 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.0 | 0.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0052692 | raffinose alpha-galactosidase activity(GO:0052692) |
| 0.0 | 0.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 3.0 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 4.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.3 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0035870 | dITP diphosphatase activity(GO:0035870) |
| 0.0 | 0.2 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 1.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.3 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 1.0 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.0 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.0 | 1.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 0.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.9 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 4.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.9 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.7 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.0 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.8 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 4.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.9 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |