Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
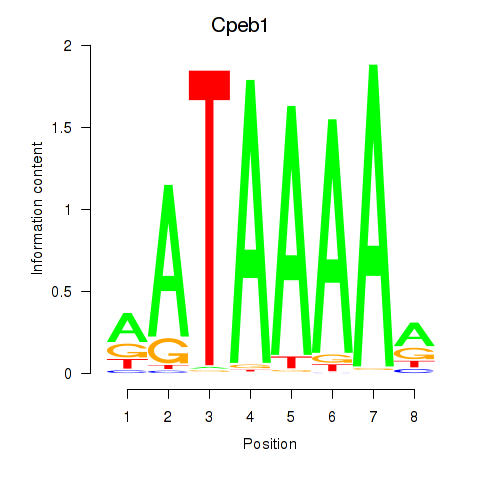
Results for Cpeb1
Z-value: 0.99
Transcription factors associated with Cpeb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cpeb1
|
ENSMUSG00000025586.18 | cytoplasmic polyadenylation element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cpeb1 | mm39_v1_chr7_-_81104423_81104512 | -0.47 | 3.9e-03 | Click! |
Activity profile of Cpeb1 motif
Sorted Z-values of Cpeb1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_103502404 | 6.58 |
ENSMUST00000033229.5
|
Hbb-y
|
hemoglobin Y, beta-like embryonic chain |
| chr9_+_96141299 | 6.47 |
ENSMUST00000179065.8
|
Tfdp2
|
transcription factor Dp 2 |
| chr9_+_96141317 | 6.38 |
ENSMUST00000165768.4
|
Tfdp2
|
transcription factor Dp 2 |
| chr5_+_90638580 | 6.07 |
ENSMUST00000042755.7
ENSMUST00000200693.2 |
Afp
|
alpha fetoprotein |
| chr4_-_87951565 | 6.02 |
ENSMUST00000078090.12
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr7_-_103463120 | 4.46 |
ENSMUST00000098192.4
|
Hbb-bt
|
hemoglobin, beta adult t chain |
| chr7_-_103477126 | 4.38 |
ENSMUST00000023934.8
|
Hbb-bs
|
hemoglobin, beta adult s chain |
| chr15_-_36609208 | 4.20 |
ENSMUST00000001809.15
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr7_-_16790594 | 4.11 |
ENSMUST00000037762.11
|
Hif3a
|
hypoxia inducible factor 3, alpha subunit |
| chr11_-_98915005 | 4.07 |
ENSMUST00000068031.8
|
Top2a
|
topoisomerase (DNA) II alpha |
| chr14_-_70873385 | 3.93 |
ENSMUST00000228295.2
ENSMUST00000022695.16 |
Dmtn
|
dematin actin binding protein |
| chr5_+_90920353 | 3.93 |
ENSMUST00000202625.2
|
Pf4
|
platelet factor 4 |
| chr7_-_103492361 | 3.89 |
ENSMUST00000063957.6
|
Hbb-bh1
|
hemoglobin Z, beta-like embryonic chain |
| chr5_+_90920294 | 3.74 |
ENSMUST00000031320.8
|
Pf4
|
platelet factor 4 |
| chr5_-_73413888 | 3.60 |
ENSMUST00000101127.12
|
Fryl
|
FRY like transcription coactivator |
| chr16_+_36097313 | 3.38 |
ENSMUST00000232150.2
|
Stfa1
|
stefin A1 |
| chr17_+_41121979 | 3.32 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr5_+_76988444 | 3.30 |
ENSMUST00000120639.9
ENSMUST00000163347.8 ENSMUST00000121851.2 |
Cracd
|
capping protein inhibiting regulator of actin |
| chr9_+_96140781 | 3.18 |
ENSMUST00000190104.7
ENSMUST00000179416.8 ENSMUST00000189606.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr1_-_45542442 | 3.10 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2 |
| chr12_-_55033130 | 3.09 |
ENSMUST00000173433.8
ENSMUST00000173803.2 |
Baz1a
Gm20403
|
bromodomain adjacent to zinc finger domain 1A predicted gene 20403 |
| chr8_-_73059104 | 3.06 |
ENSMUST00000075602.8
|
Gm10282
|
predicted pseudogene 10282 |
| chr12_-_55033113 | 3.00 |
ENSMUST00000038926.13
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr7_+_89780785 | 2.96 |
ENSMUST00000208684.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr2_+_155453103 | 2.88 |
ENSMUST00000092995.6
|
Myh7b
|
myosin, heavy chain 7B, cardiac muscle, beta |
| chr10_+_115653152 | 2.75 |
ENSMUST00000080630.11
ENSMUST00000179196.3 ENSMUST00000035563.15 |
Tspan8
|
tetraspanin 8 |
| chr11_+_31822211 | 2.73 |
ENSMUST00000020543.13
ENSMUST00000109412.9 |
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chrX_+_149330371 | 2.65 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr10_-_35587888 | 2.43 |
ENSMUST00000080898.4
|
Amd2
|
S-adenosylmethionine decarboxylase 2 |
| chr3_-_106126794 | 2.43 |
ENSMUST00000082219.6
|
Chil4
|
chitinase-like 4 |
| chr6_-_7693110 | 2.43 |
ENSMUST00000126303.8
|
Asns
|
asparagine synthetase |
| chr9_+_65797519 | 2.34 |
ENSMUST00000045802.7
|
Pclaf
|
PCNA clamp associated factor |
| chr9_+_108437485 | 2.26 |
ENSMUST00000081111.14
ENSMUST00000193421.2 |
Impdh2
|
inosine monophosphate dehydrogenase 2 |
| chr10_-_40178182 | 2.22 |
ENSMUST00000099945.6
ENSMUST00000238953.2 ENSMUST00000238969.2 |
Amd1
|
S-adenosylmethionine decarboxylase 1 |
| chr10_+_53472853 | 2.21 |
ENSMUST00000219271.2
|
Asf1a
|
anti-silencing function 1A histone chaperone |
| chr10_+_45453907 | 2.21 |
ENSMUST00000037044.13
|
Hace1
|
HECT domain and ankyrin repeat containing, E3 ubiquitin protein ligase 1 |
| chrX_-_133483849 | 2.11 |
ENSMUST00000113213.2
ENSMUST00000033617.13 |
Btk
|
Bruton agammaglobulinemia tyrosine kinase |
| chr9_+_96140750 | 2.03 |
ENSMUST00000186609.7
|
Tfdp2
|
transcription factor Dp 2 |
| chr11_-_99328969 | 2.03 |
ENSMUST00000017743.3
|
Krt20
|
keratin 20 |
| chr15_+_103148824 | 2.02 |
ENSMUST00000036004.16
ENSMUST00000087351.9 ENSMUST00000231141.2 |
Hnrnpa1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr10_+_53473032 | 2.01 |
ENSMUST00000020004.8
|
Asf1a
|
anti-silencing function 1A histone chaperone |
| chr19_+_53298906 | 1.92 |
ENSMUST00000003870.15
|
Mxi1
|
MAX interactor 1, dimerization protein |
| chr14_+_26722319 | 1.89 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr14_+_66043281 | 1.83 |
ENSMUST00000022612.10
|
Pbk
|
PDZ binding kinase |
| chr17_-_33879224 | 1.75 |
ENSMUST00000130946.8
|
Hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr2_-_152672185 | 1.72 |
ENSMUST00000140436.2
|
Bcl2l1
|
BCL2-like 1 |
| chr8_+_108271643 | 1.71 |
ENSMUST00000212543.2
|
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr2_-_102731691 | 1.69 |
ENSMUST00000111192.3
ENSMUST00000111190.9 ENSMUST00000111198.9 ENSMUST00000111191.9 ENSMUST00000060516.14 ENSMUST00000099673.9 ENSMUST00000005218.15 ENSMUST00000111194.8 |
Cd44
|
CD44 antigen |
| chr2_-_70655997 | 1.64 |
ENSMUST00000038584.9
|
Tlk1
|
tousled-like kinase 1 |
| chr6_-_51443602 | 1.62 |
ENSMUST00000203253.2
|
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr13_+_44994167 | 1.62 |
ENSMUST00000173906.3
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr7_+_142052569 | 1.61 |
ENSMUST00000078497.15
ENSMUST00000105953.10 ENSMUST00000179658.8 ENSMUST00000105954.10 ENSMUST00000105952.10 ENSMUST00000105955.8 ENSMUST00000074187.13 ENSMUST00000169299.9 ENSMUST00000105957.10 ENSMUST00000180152.8 ENSMUST00000105950.11 ENSMUST00000105958.10 ENSMUST00000105949.8 |
Tnnt3
|
troponin T3, skeletal, fast |
| chr1_-_82746169 | 1.60 |
ENSMUST00000027331.3
|
Tm4sf20
|
transmembrane 4 L six family member 20 |
| chr4_-_117354249 | 1.56 |
ENSMUST00000030439.15
|
Rnf220
|
ring finger protein 220 |
| chr11_+_11634967 | 1.53 |
ENSMUST00000141436.8
ENSMUST00000126058.8 |
Ikzf1
|
IKAROS family zinc finger 1 |
| chr14_-_99231754 | 1.51 |
ENSMUST00000081987.5
|
Rpl36a-ps1
|
ribosomal protein L36A, pseudogene 1 |
| chr19_-_60770628 | 1.49 |
ENSMUST00000238125.2
|
Eif3a
|
eukaryotic translation initiation factor 3, subunit A |
| chr13_-_96803216 | 1.49 |
ENSMUST00000170287.8
|
Hmgcr
|
3-hydroxy-3-methylglutaryl-Coenzyme A reductase |
| chr3_+_113824181 | 1.47 |
ENSMUST00000123619.8
ENSMUST00000092155.12 |
Col11a1
|
collagen, type XI, alpha 1 |
| chrX_+_55825033 | 1.46 |
ENSMUST00000114772.9
ENSMUST00000114768.10 ENSMUST00000155882.8 |
Fhl1
|
four and a half LIM domains 1 |
| chr18_-_77855446 | 1.45 |
ENSMUST00000048192.9
|
Haus1
|
HAUS augmin-like complex, subunit 1 |
| chr4_+_140428777 | 1.44 |
ENSMUST00000138808.8
ENSMUST00000038893.6 |
Rcc2
|
regulator of chromosome condensation 2 |
| chr1_+_134109888 | 1.42 |
ENSMUST00000156873.8
|
Chil1
|
chitinase-like 1 |
| chr9_+_54606832 | 1.39 |
ENSMUST00000070070.8
|
Dnaja4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr10_-_30647836 | 1.38 |
ENSMUST00000215926.2
ENSMUST00000213836.2 |
Ncoa7
|
nuclear receptor coactivator 7 |
| chr6_-_87958611 | 1.37 |
ENSMUST00000056403.7
|
H1f10
|
H1.10 linker histone |
| chr6_+_116327853 | 1.37 |
ENSMUST00000140884.8
ENSMUST00000129170.8 |
Marchf8
|
membrane associated ring-CH-type finger 8 |
| chrX_+_55824797 | 1.35 |
ENSMUST00000114773.10
|
Fhl1
|
four and a half LIM domains 1 |
| chr1_-_128520002 | 1.34 |
ENSMUST00000052172.7
ENSMUST00000142893.2 |
Cxcr4
|
chemokine (C-X-C motif) receptor 4 |
| chrX_+_141608694 | 1.34 |
ENSMUST00000112888.2
|
Tmem164
|
transmembrane protein 164 |
| chr7_-_115445352 | 1.33 |
ENSMUST00000206369.2
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr2_+_13578738 | 1.31 |
ENSMUST00000141365.3
ENSMUST00000028062.8 |
Vim
|
vimentin |
| chr14_-_55344004 | 1.31 |
ENSMUST00000036041.15
|
Ap1g2
|
adaptor protein complex AP-1, gamma 2 subunit |
| chr9_+_54606798 | 1.30 |
ENSMUST00000154690.8
|
Dnaja4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr4_-_136329953 | 1.30 |
ENSMUST00000105847.8
ENSMUST00000116273.9 |
Kdm1a
|
lysine (K)-specific demethylase 1A |
| chr1_-_66984178 | 1.30 |
ENSMUST00000027151.12
|
Myl1
|
myosin, light polypeptide 1 |
| chr2_+_13579092 | 1.28 |
ENSMUST00000193675.2
|
Vim
|
vimentin |
| chr9_-_71803354 | 1.27 |
ENSMUST00000184448.8
|
Tcf12
|
transcription factor 12 |
| chr2_-_5838489 | 1.25 |
ENSMUST00000128467.4
|
Cdc123
|
cell division cycle 123 |
| chr5_+_86219593 | 1.24 |
ENSMUST00000198435.5
ENSMUST00000031171.9 |
Stap1
|
signal transducing adaptor family member 1 |
| chr15_+_61857226 | 1.24 |
ENSMUST00000161976.8
ENSMUST00000022971.8 |
Myc
|
myelocytomatosis oncogene |
| chr10_-_30076543 | 1.23 |
ENSMUST00000099985.6
|
Cenpw
|
centromere protein W |
| chr13_+_37529184 | 1.23 |
ENSMUST00000021860.7
|
Ly86
|
lymphocyte antigen 86 |
| chr3_+_60503051 | 1.23 |
ENSMUST00000192757.6
ENSMUST00000193518.6 ENSMUST00000195817.3 |
Mbnl1
|
muscleblind like splicing factor 1 |
| chr3_+_96177010 | 1.22 |
ENSMUST00000051089.4
ENSMUST00000177113.2 |
Gm42743
H2bc18
|
predicted gene 42743 H2B clustered histone 18 |
| chr10_-_127025851 | 1.22 |
ENSMUST00000222006.2
ENSMUST00000019611.15 ENSMUST00000219245.2 |
Arhgef25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chr4_+_11758147 | 1.22 |
ENSMUST00000029871.12
ENSMUST00000108303.2 |
Cdh17
|
cadherin 17 |
| chr11_-_95896721 | 1.21 |
ENSMUST00000013559.3
|
Igf2bp1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr19_+_39049442 | 1.21 |
ENSMUST00000087236.5
|
Cyp2c65
|
cytochrome P450, family 2, subfamily c, polypeptide 65 |
| chr15_-_101389384 | 1.20 |
ENSMUST00000023718.9
|
Krt83
|
keratin 83 |
| chr15_+_61857390 | 1.20 |
ENSMUST00000159327.2
ENSMUST00000167731.8 |
Myc
|
myelocytomatosis oncogene |
| chr10_-_37014859 | 1.19 |
ENSMUST00000092584.6
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr7_-_121620366 | 1.19 |
ENSMUST00000033160.15
|
Gga2
|
golgi associated, gamma adaptin ear containing, ARF binding protein 2 |
| chr3_-_14843512 | 1.18 |
ENSMUST00000094365.11
|
Car1
|
carbonic anhydrase 1 |
| chr2_-_151586063 | 1.17 |
ENSMUST00000109869.2
|
Psmf1
|
proteasome (prosome, macropain) inhibitor subunit 1 |
| chr13_+_93441447 | 1.15 |
ENSMUST00000109497.8
ENSMUST00000109498.8 ENSMUST00000060490.11 ENSMUST00000109492.9 ENSMUST00000109496.8 ENSMUST00000109495.8 |
Homer1
|
homer scaffolding protein 1 |
| chr8_+_85449632 | 1.15 |
ENSMUST00000098571.5
|
G430095P16Rik
|
RIKEN cDNA G430095P16 gene |
| chr13_+_94954202 | 1.13 |
ENSMUST00000220825.2
|
Tbca
|
tubulin cofactor A |
| chr4_-_44168339 | 1.11 |
ENSMUST00000045793.15
|
Rnf38
|
ring finger protein 38 |
| chr12_-_36092475 | 1.11 |
ENSMUST00000020896.17
|
Tspan13
|
tetraspanin 13 |
| chr1_-_66974492 | 1.11 |
ENSMUST00000120415.8
ENSMUST00000119429.8 |
Myl1
|
myosin, light polypeptide 1 |
| chr17_+_18108086 | 1.10 |
ENSMUST00000149944.2
|
Fpr2
|
formyl peptide receptor 2 |
| chr13_+_22220000 | 1.09 |
ENSMUST00000110455.4
|
H2bc12
|
H2B clustered histone 12 |
| chr7_-_115445315 | 1.08 |
ENSMUST00000166207.3
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr14_+_79753055 | 1.08 |
ENSMUST00000110835.3
ENSMUST00000227192.2 |
Elf1
|
E74-like factor 1 |
| chr12_-_57244096 | 1.05 |
ENSMUST00000044634.12
|
Slc25a21
|
solute carrier family 25 (mitochondrial oxodicarboxylate carrier), member 21 |
| chr6_+_8520006 | 1.05 |
ENSMUST00000162567.8
ENSMUST00000161217.8 |
Glcci1
|
glucocorticoid induced transcript 1 |
| chr17_-_10501816 | 1.05 |
ENSMUST00000233684.2
|
Qk
|
quaking, KH domain containing RNA binding |
| chrX_+_133618693 | 1.03 |
ENSMUST00000113201.8
ENSMUST00000051256.10 ENSMUST00000113199.8 ENSMUST00000035748.14 ENSMUST00000113198.8 ENSMUST00000113197.2 |
Armcx1
|
armadillo repeat containing, X-linked 1 |
| chr6_+_125090473 | 1.02 |
ENSMUST00000124317.2
|
Chd4
|
chromodomain helicase DNA binding protein 4 |
| chr6_+_5390386 | 1.01 |
ENSMUST00000183358.2
|
Asb4
|
ankyrin repeat and SOCS box-containing 4 |
| chr11_-_106084334 | 1.01 |
ENSMUST00000007444.14
ENSMUST00000152008.2 ENSMUST00000103072.10 ENSMUST00000106867.2 |
Strada
|
STE20-related kinase adaptor alpha |
| chr13_-_23806530 | 0.99 |
ENSMUST00000062045.4
|
H1f4
|
H1.4 linker histone, cluster member |
| chr13_-_103901010 | 0.98 |
ENSMUST00000210489.2
|
Srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr13_+_83652352 | 0.98 |
ENSMUST00000198916.5
ENSMUST00000200123.5 ENSMUST00000005722.14 ENSMUST00000163888.8 |
Mef2c
|
myocyte enhancer factor 2C |
| chr2_+_84818538 | 0.97 |
ENSMUST00000028466.12
|
Prg3
|
proteoglycan 3 |
| chr6_-_16898440 | 0.96 |
ENSMUST00000031533.11
|
Tfec
|
transcription factor EC |
| chr6_-_97182509 | 0.96 |
ENSMUST00000164744.8
ENSMUST00000089287.7 |
Uba3
|
ubiquitin-like modifier activating enzyme 3 |
| chr15_-_66673425 | 0.95 |
ENSMUST00000168589.8
|
Sla
|
src-like adaptor |
| chr17_+_18108102 | 0.94 |
ENSMUST00000054871.12
ENSMUST00000064068.5 |
Fpr3
Fpr2
|
formyl peptide receptor 3 formyl peptide receptor 2 |
| chr4_-_133695204 | 0.94 |
ENSMUST00000100472.10
ENSMUST00000136327.2 |
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr1_+_135727140 | 0.94 |
ENSMUST00000152208.8
ENSMUST00000152075.8 |
Tnni1
|
troponin I, skeletal, slow 1 |
| chr2_-_148285450 | 0.93 |
ENSMUST00000099269.4
|
Cd93
|
CD93 antigen |
| chr3_-_88317601 | 0.92 |
ENSMUST00000193338.6
ENSMUST00000056370.13 |
Pmf1
|
polyamine-modulated factor 1 |
| chr3_+_5815863 | 0.92 |
ENSMUST00000192045.2
|
Gm8797
|
predicted pseudogene 8797 |
| chr1_-_79838897 | 0.91 |
ENSMUST00000190724.2
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr11_+_120123727 | 0.90 |
ENSMUST00000122148.8
ENSMUST00000044985.14 |
Bahcc1
|
BAH domain and coiled-coil containing 1 |
| chrX_-_74460168 | 0.90 |
ENSMUST00000033543.14
ENSMUST00000149863.3 ENSMUST00000114081.2 |
Cmc4
Mtcp1
|
C-x(9)-C motif containing 4 mature T cell proliferation 1 |
| chr19_+_55730696 | 0.88 |
ENSMUST00000153888.9
ENSMUST00000127233.9 ENSMUST00000061496.17 ENSMUST00000111656.8 ENSMUST00000111657.11 |
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr10_-_30531832 | 0.88 |
ENSMUST00000217138.2
ENSMUST00000217644.2 ENSMUST00000216172.2 |
Ncoa7
|
nuclear receptor coactivator 7 |
| chr16_+_94370869 | 0.87 |
ENSMUST00000119878.8
|
Dyrk1a
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1a |
| chr1_+_135727228 | 0.87 |
ENSMUST00000154463.8
ENSMUST00000139986.8 |
Tnni1
|
troponin I, skeletal, slow 1 |
| chr1_+_177272215 | 0.86 |
ENSMUST00000192851.2
ENSMUST00000193480.2 ENSMUST00000195388.2 |
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr1_-_66984521 | 0.86 |
ENSMUST00000160100.2
|
Myl1
|
myosin, light polypeptide 1 |
| chr11_-_102210568 | 0.85 |
ENSMUST00000173870.8
|
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr17_+_28059129 | 0.84 |
ENSMUST00000233657.2
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr1_-_66974694 | 0.83 |
ENSMUST00000186202.7
|
Myl1
|
myosin, light polypeptide 1 |
| chr4_-_133981387 | 0.82 |
ENSMUST00000060050.6
|
Grrp1
|
glycine/arginine rich protein 1 |
| chr11_-_82799186 | 0.82 |
ENSMUST00000103213.10
|
Nle1
|
notchless homolog 1 |
| chr3_+_68598757 | 0.82 |
ENSMUST00000107816.4
|
Il12a
|
interleukin 12a |
| chr5_-_36771074 | 0.82 |
ENSMUST00000132383.6
ENSMUST00000174019.2 |
D5Ertd579e
Gm42936
|
DNA segment, Chr 5, ERATO Doi 579, expressed predicted gene 42936 |
| chr9_+_108673171 | 0.81 |
ENSMUST00000195514.6
ENSMUST00000085018.6 ENSMUST00000192028.6 |
Ip6k2
|
inositol hexaphosphate kinase 2 |
| chr19_+_39102342 | 0.80 |
ENSMUST00000087234.3
|
Cyp2c66
|
cytochrome P450, family 2, subfamily c, polypeptide 66 |
| chr2_+_90817948 | 0.79 |
ENSMUST00000111452.8
ENSMUST00000111455.9 |
Celf1
|
CUGBP, Elav-like family member 1 |
| chr15_+_54274151 | 0.79 |
ENSMUST00000036737.4
|
Colec10
|
collectin sub-family member 10 |
| chr5_-_107437427 | 0.79 |
ENSMUST00000031224.15
|
Tgfbr3
|
transforming growth factor, beta receptor III |
| chr7_-_92319126 | 0.78 |
ENSMUST00000119954.9
|
Pcf11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr9_-_85209162 | 0.78 |
ENSMUST00000034802.15
|
Tent5a
|
terminal nucleotidyltransferase 5A |
| chr10_-_30647881 | 0.78 |
ENSMUST00000215740.2
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr6_+_149226891 | 0.78 |
ENSMUST00000189837.2
|
Resf1
|
retroelement silencing factor 1 |
| chr8_-_61407760 | 0.78 |
ENSMUST00000110302.8
|
Clcn3
|
chloride channel, voltage-sensitive 3 |
| chr10_-_30531768 | 0.78 |
ENSMUST00000092610.12
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr18_+_21205386 | 0.78 |
ENSMUST00000082235.5
|
Mep1b
|
meprin 1 beta |
| chr2_+_36120438 | 0.77 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr13_+_49735938 | 0.77 |
ENSMUST00000221170.2
|
Omd
|
osteomodulin |
| chrX_-_42363663 | 0.76 |
ENSMUST00000016294.8
|
Tenm1
|
teneurin transmembrane protein 1 |
| chr9_-_85209340 | 0.76 |
ENSMUST00000187711.2
|
Tent5a
|
terminal nucleotidyltransferase 5A |
| chr6_-_47571901 | 0.76 |
ENSMUST00000081721.13
ENSMUST00000114618.8 ENSMUST00000114616.8 |
Ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr8_-_58106057 | 0.75 |
ENSMUST00000034021.12
|
Galnt7
|
polypeptide N-acetylgalactosaminyltransferase 7 |
| chr17_+_28059099 | 0.75 |
ENSMUST00000233752.2
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr11_-_120489172 | 0.75 |
ENSMUST00000026125.3
|
Alyref
|
Aly/REF export factor |
| chr10_-_129738595 | 0.74 |
ENSMUST00000071557.2
|
Olfr815
|
olfactory receptor 815 |
| chr11_-_78074377 | 0.74 |
ENSMUST00000102483.5
|
Rpl23a
|
ribosomal protein L23A |
| chr9_+_122752116 | 0.74 |
ENSMUST00000051667.14
|
Zfp105
|
zinc finger protein 105 |
| chr15_-_98729333 | 0.73 |
ENSMUST00000168846.3
|
Prkag1
|
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr2_-_51039112 | 0.72 |
ENSMUST00000154545.2
ENSMUST00000017288.9 |
Rnd3
|
Rho family GTPase 3 |
| chrX_+_95498965 | 0.72 |
ENSMUST00000033553.14
|
Heph
|
hephaestin |
| chr9_+_56982622 | 0.72 |
ENSMUST00000167715.8
|
Sin3a
|
transcriptional regulator, SIN3A (yeast) |
| chr17_+_24026892 | 0.71 |
ENSMUST00000191385.3
|
Srrm2
|
serine/arginine repetitive matrix 2 |
| chr4_-_44168252 | 0.71 |
ENSMUST00000145760.8
ENSMUST00000128426.8 |
Rnf38
|
ring finger protein 38 |
| chr17_-_78991691 | 0.71 |
ENSMUST00000145480.2
|
Strn
|
striatin, calmodulin binding protein |
| chr13_+_104424359 | 0.71 |
ENSMUST00000065766.7
|
Adamts6
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 6 |
| chr13_-_29137673 | 0.71 |
ENSMUST00000067230.6
|
Sox4
|
SRY (sex determining region Y)-box 4 |
| chr3_-_92616380 | 0.70 |
ENSMUST00000090867.2
|
Lce1e
|
late cornified envelope 1E |
| chr13_+_13612136 | 0.70 |
ENSMUST00000005532.9
|
Nid1
|
nidogen 1 |
| chr6_-_129252396 | 0.70 |
ENSMUST00000032259.6
|
Cd69
|
CD69 antigen |
| chr17_+_35278011 | 0.70 |
ENSMUST00000007255.13
ENSMUST00000174493.8 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr11_+_29080733 | 0.69 |
ENSMUST00000020756.9
|
Pnpt1
|
polyribonucleotide nucleotidyltransferase 1 |
| chrX_+_158623460 | 0.69 |
ENSMUST00000112451.8
ENSMUST00000112453.9 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr3_-_58792633 | 0.69 |
ENSMUST00000055636.13
ENSMUST00000072551.7 ENSMUST00000051408.8 |
Clrn1
|
clarin 1 |
| chr10_-_99595498 | 0.69 |
ENSMUST00000056085.6
|
Csl
|
citrate synthase like |
| chr11_+_46701619 | 0.69 |
ENSMUST00000068877.7
|
Timd4
|
T cell immunoglobulin and mucin domain containing 4 |
| chr11_+_104468107 | 0.69 |
ENSMUST00000106956.10
|
Myl4
|
myosin, light polypeptide 4 |
| chr1_+_135727571 | 0.69 |
ENSMUST00000148201.8
|
Tnni1
|
troponin I, skeletal, slow 1 |
| chrX_-_12539778 | 0.68 |
ENSMUST00000060108.7
|
1810030O07Rik
|
RIKEN cDNA 1810030O07 gene |
| chr17_+_36179273 | 0.67 |
ENSMUST00000190496.2
|
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr17_+_28059036 | 0.66 |
ENSMUST00000071006.9
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr9_-_114219685 | 0.66 |
ENSMUST00000084881.5
|
Crtap
|
cartilage associated protein |
| chr10_-_88440869 | 0.66 |
ENSMUST00000119185.8
ENSMUST00000238199.2 |
Mybpc1
|
myosin binding protein C, slow-type |
| chr1_+_128079543 | 0.66 |
ENSMUST00000189317.3
|
R3hdm1
|
R3H domain containing 1 |
| chr5_+_115697526 | 0.66 |
ENSMUST00000086519.12
ENSMUST00000156359.2 ENSMUST00000152976.2 |
Rplp0
|
ribosomal protein, large, P0 |
| chr5_+_107645626 | 0.66 |
ENSMUST00000152474.8
ENSMUST00000060553.8 |
Btbd8
|
BTB (POZ) domain containing 8 |
| chr6_-_136781406 | 0.65 |
ENSMUST00000179285.3
|
H4f16
|
H4 histone 16 |
| chr2_+_62494622 | 0.65 |
ENSMUST00000028257.3
|
Gca
|
grancalcin |
| chr13_-_22219738 | 0.65 |
ENSMUST00000091742.6
|
H2ac12
|
H2A clustered histone 12 |
| chr4_+_108336164 | 0.65 |
ENSMUST00000155068.2
|
Tut4
|
terminal uridylyl transferase 4 |
| chr14_+_67982630 | 0.65 |
ENSMUST00000223929.2
ENSMUST00000111095.4 |
Gnrh1
|
gonadotropin releasing hormone 1 |
| chr10_-_88440996 | 0.64 |
ENSMUST00000121629.8
|
Mybpc1
|
myosin binding protein C, slow-type |
| chr7_+_67602565 | 0.64 |
ENSMUST00000005671.10
|
Igf1r
|
insulin-like growth factor I receptor |
| chr4_-_136563154 | 0.64 |
ENSMUST00000105846.9
ENSMUST00000059287.14 ENSMUST00000105845.9 |
Ephb2
|
Eph receptor B2 |
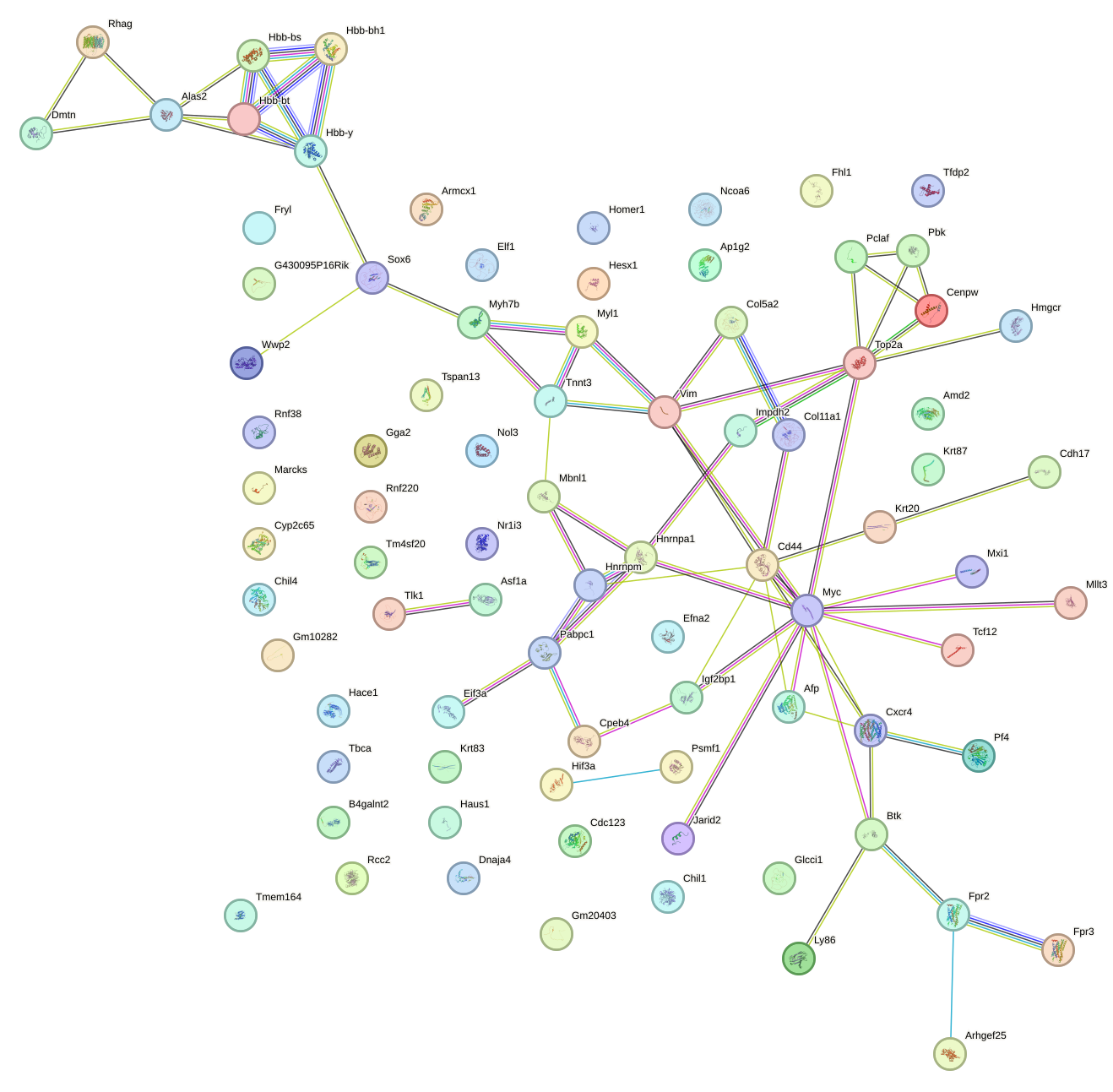
Network of associatons between targets according to the STRING database.
First level regulatory network of Cpeb1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 1.5 | 7.7 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 1.2 | 10.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.8 | 3.3 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.8 | 4.1 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.8 | 3.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.7 | 2.1 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.7 | 3.9 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.6 | 2.4 | GO:0090095 | lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.6 | 2.4 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.6 | 4.2 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.5 | 1.6 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.5 | 1.5 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 0.5 | 3.0 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.5 | 1.4 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.4 | 1.7 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.4 | 1.7 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.4 | 1.2 | GO:1903969 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.4 | 1.2 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.4 | 4.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.4 | 1.5 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.4 | 6.1 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.3 | 3.6 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.3 | 1.3 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.3 | 2.6 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.3 | 2.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.3 | 0.9 | GO:0044704 | mating plug formation(GO:0042628) single-organism reproductive behavior(GO:0044704) post-mating behavior(GO:0045297) seminal vesicle development(GO:0061107) |
| 0.3 | 5.9 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.3 | 2.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.3 | 2.5 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.3 | 1.3 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.3 | 0.8 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) |
| 0.3 | 0.8 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.3 | 0.8 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.2 | 2.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 1.5 | GO:0035989 | tendon development(GO:0035989) |
| 0.2 | 1.0 | GO:0045575 | basophil activation(GO:0045575) |
| 0.2 | 1.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 | 1.0 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.2 | 0.7 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.2 | 0.7 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.2 | 1.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.2 | 0.7 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.2 | 0.6 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.2 | 0.6 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.2 | 0.6 | GO:0061193 | taste bud development(GO:0061193) |
| 0.2 | 0.4 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.2 | 3.8 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.2 | 1.9 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.2 | 2.0 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.2 | 0.7 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.2 | 1.9 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.2 | 0.3 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.2 | 0.6 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.4 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 2.6 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 2.3 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 0.9 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.1 | 0.4 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 2.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 0.6 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.1 | 2.4 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 0.5 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.1 | 1.6 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.9 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.4 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 1.3 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) myoblast fate commitment(GO:0048625) |
| 0.1 | 0.5 | GO:0048691 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.1 | 0.7 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.1 | 0.5 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.4 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.8 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.6 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) regulation of ovarian follicle development(GO:2000354) |
| 0.1 | 1.6 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 1.2 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.6 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.1 | 0.2 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.5 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.1 | 0.6 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.1 | 0.4 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.4 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 3.9 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.3 | GO:0070948 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.1 | 0.3 | GO:1903210 | glomerular visceral epithelial cell apoptotic process(GO:1903210) regulation of glomerular visceral epithelial cell apoptotic process(GO:1904633) positive regulation of glomerular visceral epithelial cell apoptotic process(GO:1904635) positive regulation of progesterone biosynthetic process(GO:2000184) |
| 0.1 | 0.8 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.1 | 0.8 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.1 | 0.4 | GO:0045186 | zonula adherens assembly(GO:0045186) |
| 0.1 | 1.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 1.0 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.7 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.5 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.1 | 0.8 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.1 | 1.0 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.1 | 0.5 | GO:0055099 | regulation of Cdc42 protein signal transduction(GO:0032489) response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 1.5 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 1.7 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 1.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 7.5 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.1 | 0.5 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.6 | GO:0061591 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.1 | 0.5 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.1 | 0.2 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.1 | 0.1 | GO:0042488 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) |
| 0.1 | 0.1 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.1 | 0.8 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.1 | 0.7 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.7 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 1.5 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 0.5 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.1 | 0.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.4 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 0.1 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.1 | 0.7 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 2.8 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 0.1 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 1.4 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 1.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.3 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 0.5 | GO:0007549 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 | 0.4 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.1 | 0.4 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.1 | 0.3 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.1 | 0.5 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 1.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 2.8 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.5 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.3 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.3 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.3 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.8 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.4 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.4 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.0 | GO:0046101 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.2 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.5 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.2 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 1.2 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.2 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.2 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 0.2 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.1 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.6 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.3 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 1.8 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 1.1 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 4.9 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 1.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.0 | 0.7 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.1 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.4 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.0 | 0.3 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.0 | 1.4 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 2.8 | GO:1901799 | negative regulation of proteasomal protein catabolic process(GO:1901799) |
| 0.0 | 0.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.8 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.2 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.1 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.0 | 0.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 1.0 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.3 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.5 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.2 | GO:0060313 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.0 | 0.1 | GO:1900190 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.0 | 1.7 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0060685 | regulation of prostatic bud formation(GO:0060685) negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.1 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.0 | 0.3 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 3.9 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.2 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.5 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 1.4 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.3 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.7 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.2 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.4 | GO:0098534 | centriole replication(GO:0007099) centriole assembly(GO:0098534) |
| 0.0 | 3.4 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.0 | 0.5 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.0 | 0.4 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:0002849 | positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.0 | 0.1 | GO:0051025 | negative regulation of immunoglobulin secretion(GO:0051025) |
| 0.0 | 1.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.0 | 0.7 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 1.0 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 14.3 | GO:0000278 | mitotic cell cycle(GO:0000278) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.5 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.2 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.5 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.2 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 1.8 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.0 | 0.3 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 1.1 | GO:0051302 | regulation of cell division(GO:0051302) |
| 0.0 | 0.5 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0045901 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.2 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.1 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 1.1 | GO:0042303 | molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.0 | 0.2 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.1 | GO:1990000 | amyloid fibril formation(GO:1990000) |
| 0.0 | 0.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 19.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 1.5 | 6.1 | GO:0008623 | CHRAC(GO:0008623) |
| 1.0 | 3.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.6 | 3.9 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.5 | 7.7 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.5 | 1.5 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.4 | 3.0 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.3 | 0.8 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.3 | 6.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 1.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 2.9 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 4.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 1.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.2 | 0.9 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.2 | 1.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 1.6 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.2 | 1.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 0.5 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 2.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 1.5 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 1.7 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.5 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.6 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 1.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 21.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 6.0 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 0.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 2.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 0.3 | GO:1990843 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.1 | 0.3 | GO:0032998 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.1 | 1.5 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 0.3 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 0.6 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 0.8 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 0.6 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 0.3 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 2.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 2.4 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 5.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.4 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 0.9 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 0.5 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 2.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 1.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.7 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.4 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.7 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.0 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 2.3 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.3 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 3.7 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.8 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 1.0 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.6 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.0 | 0.8 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.1 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 4.2 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 1.0 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.4 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.5 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.4 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:0016011 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.4 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 3.1 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 2.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 4.0 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 4.4 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.7 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 6.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.8 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.0 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 1.1 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.6 | GO:0045171 | intercellular bridge(GO:0045171) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 19.3 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 1.5 | 6.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 1.3 | 7.7 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 1.0 | 4.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.9 | 2.6 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.7 | 2.0 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.7 | 4.6 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.6 | 2.4 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.6 | 2.2 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.5 | 1.5 | GO:0042282 | hydroxymethylglutaryl-CoA reductase (NADPH) activity(GO:0004420) hydroxymethylglutaryl-CoA reductase activity(GO:0042282) |
| 0.4 | 2.6 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.4 | 1.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.4 | 2.0 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.4 | 2.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.3 | 4.0 | GO:0008061 | chitin binding(GO:0008061) |
| 0.3 | 1.2 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.3 | 1.3 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.2 | 2.5 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.2 | 1.6 | GO:0031013 | troponin C binding(GO:0030172) calcium-dependent ATPase activity(GO:0030899) troponin I binding(GO:0031013) |
| 0.2 | 2.8 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 0.6 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.2 | 0.6 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.2 | 3.0 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 1.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.2 | 0.6 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.2 | 1.7 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 2.2 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.2 | 6.4 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.2 | 0.5 | GO:0034188 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.2 | 0.8 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.2 | 3.3 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.1 | 0.4 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.1 | 0.4 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 2.6 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.8 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.1 | 0.8 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.1 | 0.5 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 1.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 1.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 1.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 0.9 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.7 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 1.7 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.1 | 0.7 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.5 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 2.0 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.3 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 3.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.7 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 1.6 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.8 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.2 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 0.3 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.7 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 1.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.7 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 1.0 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 0.4 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 1.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.3 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.1 | 0.9 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.5 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.3 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.1 | 0.4 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 4.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.3 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 3.0 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.5 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 1.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 4.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.7 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.5 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 27.3 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 1.2 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 1.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.4 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 1.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 2.7 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.8 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 1.7 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.5 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.9 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 4.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.6 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 1.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 3.2 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.1 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 1.3 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 1.5 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.2 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 2.0 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 1.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 1.3 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.0 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.2 | 4.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 20.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 1.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 8.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 1.7 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 4.3 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 1.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 4.9 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 2.1 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 3.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 2.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 2.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.6 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.4 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 1.3 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 3.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.1 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 1.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.8 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.7 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.9 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.7 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.7 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 1.3 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.2 | 11.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 4.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 1.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 2.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 2.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.7 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 2.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 3.1 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 5.4 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 1.7 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 2.4 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.1 | 3.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 4.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 0.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 3.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.0 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.5 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 5.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 1.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 1.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.7 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 3.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 3.4 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 1.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.4 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |