Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
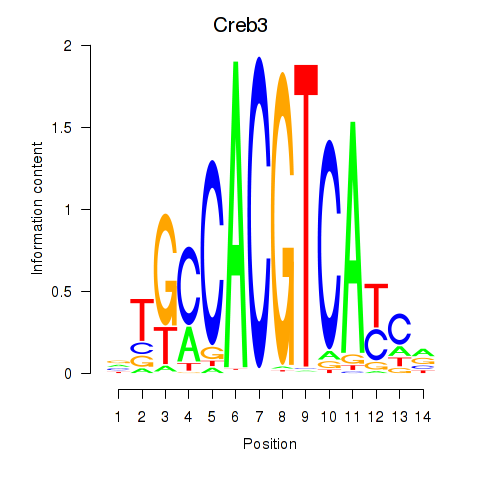
Results for Creb3
Z-value: 1.09
Transcription factors associated with Creb3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Creb3
|
ENSMUSG00000028466.16 | cAMP responsive element binding protein 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Creb3 | mm39_v1_chr4_+_43562706_43562947 | 0.36 | 3.0e-02 | Click! |
Activity profile of Creb3 motif
Sorted Z-values of Creb3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_58152295 | 8.17 |
ENSMUST00000040999.14
ENSMUST00000162011.3 |
Aox3
|
aldehyde oxidase 3 |
| chr13_-_4573312 | 7.68 |
ENSMUST00000221564.2
ENSMUST00000078239.5 ENSMUST00000080361.13 |
Akr1c20
|
aldo-keto reductase family 1, member C20 |
| chr17_+_35658131 | 7.07 |
ENSMUST00000071951.14
ENSMUST00000116598.10 ENSMUST00000078205.14 ENSMUST00000076256.8 |
H2-Q7
|
histocompatibility 2, Q region locus 7 |
| chr2_+_172994841 | 6.35 |
ENSMUST00000029017.6
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr3_+_123061094 | 6.04 |
ENSMUST00000047923.12
ENSMUST00000200333.2 |
Sec24d
|
Sec24 related gene family, member D (S. cerevisiae) |
| chr7_-_34914675 | 5.88 |
ENSMUST00000118444.3
ENSMUST00000122409.8 |
Lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr9_+_103940575 | 4.68 |
ENSMUST00000120854.8
|
Acad11
|
acyl-Coenzyme A dehydrogenase family, member 11 |
| chr4_-_104967032 | 4.33 |
ENSMUST00000030243.8
|
Prkaa2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit |
| chr17_-_57535003 | 4.16 |
ENSMUST00000177046.2
ENSMUST00000024988.15 |
C3
|
complement component 3 |
| chr10_-_4382311 | 4.03 |
ENSMUST00000126102.8
ENSMUST00000131853.2 ENSMUST00000042251.11 |
Rmnd1
|
required for meiotic nuclear division 1 homolog |
| chr9_+_44290832 | 3.94 |
ENSMUST00000161318.8
ENSMUST00000217019.2 ENSMUST00000160902.8 |
Hyou1
|
hypoxia up-regulated 1 |
| chr16_+_13721016 | 3.92 |
ENSMUST00000128757.8
|
Mpv17l
|
Mpv17 transgene, kidney disease mutant-like |
| chr9_+_44290787 | 3.79 |
ENSMUST00000066601.13
|
Hyou1
|
hypoxia up-regulated 1 |
| chrX_+_72830668 | 3.78 |
ENSMUST00000002090.3
|
Ssr4
|
signal sequence receptor, delta |
| chr11_-_96807192 | 3.70 |
ENSMUST00000144731.8
ENSMUST00000127048.8 |
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr10_-_4382283 | 3.64 |
ENSMUST00000155172.8
|
Rmnd1
|
required for meiotic nuclear division 1 homolog |
| chr11_-_61745843 | 3.56 |
ENSMUST00000004920.4
|
Ulk2
|
unc-51 like kinase 2 |
| chrX_+_72830607 | 3.49 |
ENSMUST00000166518.8
|
Ssr4
|
signal sequence receptor, delta |
| chr10_+_41395410 | 3.44 |
ENSMUST00000019962.15
|
Cd164
|
CD164 antigen |
| chr2_+_155118217 | 3.38 |
ENSMUST00000029128.4
|
Map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr11_-_96807273 | 3.36 |
ENSMUST00000103152.11
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr14_-_59835285 | 3.31 |
ENSMUST00000022555.11
ENSMUST00000225839.2 ENSMUST00000056997.15 ENSMUST00000171683.3 ENSMUST00000167100.9 |
Cdadc1
|
cytidine and dCMP deaminase domain containing 1 |
| chr1_-_183150867 | 3.30 |
ENSMUST00000194543.4
|
Mia3
|
melanoma inhibitory activity 3 |
| chr2_+_71884943 | 3.24 |
ENSMUST00000028525.6
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr2_-_168584020 | 3.24 |
ENSMUST00000109177.8
|
Atp9a
|
ATPase, class II, type 9A |
| chr10_+_41395870 | 3.00 |
ENSMUST00000189300.2
|
Cd164
|
CD164 antigen |
| chr11_-_86884507 | 2.91 |
ENSMUST00000018571.5
|
Ypel2
|
yippee like 2 |
| chr11_-_96807233 | 2.80 |
ENSMUST00000130774.2
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr11_-_72686853 | 2.69 |
ENSMUST00000156294.8
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr2_-_143853122 | 2.67 |
ENSMUST00000016072.12
ENSMUST00000037875.6 |
Rrbp1
|
ribosome binding protein 1 |
| chr5_+_28276353 | 2.63 |
ENSMUST00000059155.11
|
Insig1
|
insulin induced gene 1 |
| chr10_+_128158413 | 2.58 |
ENSMUST00000219836.2
|
Cnpy2
|
canopy FGF signaling regulator 2 |
| chr5_+_113873864 | 2.55 |
ENSMUST00000065698.7
|
Ficd
|
FIC domain containing |
| chr4_-_118401185 | 2.51 |
ENSMUST00000128098.8
|
Tmem125
|
transmembrane protein 125 |
| chr11_-_60702081 | 2.51 |
ENSMUST00000018744.15
|
Shmt1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr7_+_101859542 | 2.43 |
ENSMUST00000140631.2
ENSMUST00000120879.8 ENSMUST00000146996.8 |
Pgap2
|
post-GPI attachment to proteins 2 |
| chr10_-_67120959 | 2.41 |
ENSMUST00000159002.2
ENSMUST00000077839.13 |
Nrbf2
|
nuclear receptor binding factor 2 |
| chr11_+_94544593 | 2.40 |
ENSMUST00000025278.8
|
Mrpl27
|
mitochondrial ribosomal protein L27 |
| chr11_-_72686627 | 2.38 |
ENSMUST00000079681.6
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr8_-_106863423 | 2.37 |
ENSMUST00000146940.2
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr11_+_70104736 | 2.36 |
ENSMUST00000171032.8
|
Slc16a11
|
solute carrier family 16 (monocarboxylic acid transporters), member 11 |
| chr2_+_155224105 | 2.35 |
ENSMUST00000134218.2
|
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr10_+_128158328 | 2.25 |
ENSMUST00000219037.2
ENSMUST00000026446.4 |
Cnpy2
|
canopy FGF signaling regulator 2 |
| chr7_+_26895206 | 2.23 |
ENSMUST00000179391.8
ENSMUST00000108379.8 |
BC024978
|
cDNA sequence BC024978 |
| chr11_-_51647290 | 2.22 |
ENSMUST00000109097.9
|
Sec24a
|
Sec24 related gene family, member A (S. cerevisiae) |
| chr4_+_59003121 | 2.20 |
ENSMUST00000095070.4
ENSMUST00000174664.2 |
Dnajc25
Gm20503
|
DnaJ heat shock protein family (Hsp40) member C25 predicted gene 20503 |
| chr8_-_106863521 | 2.20 |
ENSMUST00000115979.9
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr19_-_6886965 | 2.19 |
ENSMUST00000173091.2
|
Prdx5
|
peroxiredoxin 5 |
| chr8_+_91635192 | 2.15 |
ENSMUST00000211403.2
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr3_+_89366632 | 2.12 |
ENSMUST00000107410.8
|
Pmvk
|
phosphomevalonate kinase |
| chr19_-_6887361 | 2.08 |
ENSMUST00000025904.12
|
Prdx5
|
peroxiredoxin 5 |
| chr1_+_24717711 | 2.08 |
ENSMUST00000191471.7
|
Lmbrd1
|
LMBR1 domain containing 1 |
| chr12_+_51424343 | 2.03 |
ENSMUST00000219434.2
ENSMUST00000021335.7 |
Scfd1
|
Sec1 family domain containing 1 |
| chr6_-_71121347 | 1.97 |
ENSMUST00000160918.8
|
Thnsl2
|
threonine synthase-like 2 (bacterial) |
| chr6_-_108162513 | 1.96 |
ENSMUST00000167338.8
ENSMUST00000172188.2 ENSMUST00000032191.16 |
Sumf1
|
sulfatase modifying factor 1 |
| chr7_+_128125339 | 1.93 |
ENSMUST00000033136.9
|
Bag3
|
BCL2-associated athanogene 3 |
| chr9_-_44920899 | 1.89 |
ENSMUST00000102832.3
|
Cd3e
|
CD3 antigen, epsilon polypeptide |
| chr6_-_119825081 | 1.89 |
ENSMUST00000183703.8
ENSMUST00000183911.8 |
Erc1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr1_+_157334347 | 1.87 |
ENSMUST00000027881.15
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr6_-_71121324 | 1.87 |
ENSMUST00000074241.9
|
Thnsl2
|
threonine synthase-like 2 (bacterial) |
| chr9_-_20887967 | 1.86 |
ENSMUST00000214218.2
|
S1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr17_+_46694646 | 1.85 |
ENSMUST00000113481.9
ENSMUST00000138127.8 |
Zfp318
|
zinc finger protein 318 |
| chr17_-_56490887 | 1.84 |
ENSMUST00000019723.8
|
Mydgf
|
myeloid derived growth factor |
| chr1_+_157334298 | 1.83 |
ENSMUST00000086130.9
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr19_-_6886898 | 1.80 |
ENSMUST00000238095.2
|
Prdx5
|
peroxiredoxin 5 |
| chr2_-_104324035 | 1.78 |
ENSMUST00000111124.8
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr17_+_45874800 | 1.77 |
ENSMUST00000224905.2
ENSMUST00000226086.2 ENSMUST00000041353.7 |
Slc35b2
|
solute carrier family 35, member B2 |
| chr9_-_103940247 | 1.74 |
ENSMUST00000035166.12
|
Uba5
|
ubiquitin-like modifier activating enzyme 5 |
| chr7_+_44145987 | 1.72 |
ENSMUST00000107927.5
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr11_-_51647204 | 1.71 |
ENSMUST00000109092.8
ENSMUST00000064297.5 |
Sec24a
|
Sec24 related gene family, member A (S. cerevisiae) |
| chr7_-_44145830 | 1.71 |
ENSMUST00000118515.9
ENSMUST00000138328.3 ENSMUST00000239015.2 ENSMUST00000118808.9 |
Emc10
|
ER membrane protein complex subunit 10 |
| chr2_+_80145805 | 1.71 |
ENSMUST00000028392.8
|
Dnajc10
|
DnaJ heat shock protein family (Hsp40) member C10 |
| chr11_+_119989736 | 1.70 |
ENSMUST00000106223.4
ENSMUST00000185558.2 |
Ndufaf8
|
NADH:ubiquinone oxidoreductase complex assembly factor 8 |
| chr7_+_44146012 | 1.70 |
ENSMUST00000205422.2
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr2_+_67578556 | 1.69 |
ENSMUST00000180887.2
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr19_+_11943265 | 1.68 |
ENSMUST00000025590.11
|
Osbp
|
oxysterol binding protein |
| chr1_+_24717722 | 1.65 |
ENSMUST00000186096.7
|
Lmbrd1
|
LMBR1 domain containing 1 |
| chr12_+_69230931 | 1.63 |
ENSMUST00000060579.10
|
Mgat2
|
mannoside acetylglucosaminyltransferase 2 |
| chr7_-_121666486 | 1.63 |
ENSMUST00000033159.4
|
Ears2
|
glutamyl-tRNA synthetase 2, mitochondrial |
| chr1_+_75119419 | 1.56 |
ENSMUST00000097694.11
ENSMUST00000190240.7 |
Retreg2
|
reticulophagy regulator family member 2 |
| chr11_+_53241561 | 1.55 |
ENSMUST00000060945.12
|
Aff4
|
AF4/FMR2 family, member 4 |
| chr10_-_115087309 | 1.55 |
ENSMUST00000020339.10
|
Tbc1d15
|
TBC1 domain family, member 15 |
| chr19_+_8719033 | 1.52 |
ENSMUST00000176314.8
ENSMUST00000073430.14 ENSMUST00000175901.8 |
Stx5a
|
syntaxin 5A |
| chr1_-_175319842 | 1.52 |
ENSMUST00000195324.6
ENSMUST00000192227.6 ENSMUST00000194555.6 |
Rgs7
|
regulator of G protein signaling 7 |
| chr10_-_128758757 | 1.49 |
ENSMUST00000135161.2
|
Rdh5
|
retinol dehydrogenase 5 |
| chr1_+_75119809 | 1.47 |
ENSMUST00000186037.7
ENSMUST00000187901.2 |
Retreg2
|
reticulophagy regulator family member 2 |
| chr6_-_119825020 | 1.46 |
ENSMUST00000184864.8
|
Erc1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr9_+_43996236 | 1.45 |
ENSMUST00000065461.9
ENSMUST00000176416.8 |
Usp2
|
ubiquitin specific peptidase 2 |
| chr11_-_86698484 | 1.45 |
ENSMUST00000018569.14
|
Dhx40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr14_-_55880980 | 1.45 |
ENSMUST00000132338.8
|
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr19_+_8718777 | 1.44 |
ENSMUST00000176381.8
|
Stx5a
|
syntaxin 5A |
| chr10_+_121200984 | 1.43 |
ENSMUST00000040344.7
|
Gns
|
glucosamine (N-acetyl)-6-sulfatase |
| chr9_-_64928927 | 1.42 |
ENSMUST00000036615.7
|
Hacd3
|
3-hydroxyacyl-CoA dehydratase 3 |
| chr9_+_59446823 | 1.40 |
ENSMUST00000026262.8
|
Hexa
|
hexosaminidase A |
| chr11_-_20781009 | 1.40 |
ENSMUST00000047028.9
|
Lgalsl
|
lectin, galactoside binding-like |
| chr14_-_55880708 | 1.39 |
ENSMUST00000120041.8
ENSMUST00000121937.8 ENSMUST00000133707.2 ENSMUST00000002391.15 ENSMUST00000121791.8 |
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr10_-_128759817 | 1.39 |
ENSMUST00000131271.2
|
Bloc1s1
|
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr16_-_5021843 | 1.38 |
ENSMUST00000147567.2
ENSMUST00000023911.11 |
Nagpa
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase |
| chr14_-_55881257 | 1.37 |
ENSMUST00000122358.8
|
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr1_+_75119472 | 1.35 |
ENSMUST00000189650.7
|
Retreg2
|
reticulophagy regulator family member 2 |
| chrX_-_59937036 | 1.33 |
ENSMUST00000135107.4
|
Sox3
|
SRY (sex determining region Y)-box 3 |
| chr14_-_55881177 | 1.31 |
ENSMUST00000138085.2
|
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr19_+_8718837 | 1.28 |
ENSMUST00000177373.8
ENSMUST00000010254.16 |
Stx5a
|
syntaxin 5A |
| chr1_-_93373364 | 1.27 |
ENSMUST00000190321.7
ENSMUST00000042498.14 |
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr1_+_171173252 | 1.27 |
ENSMUST00000006579.5
|
Pfdn2
|
prefoldin 2 |
| chr7_+_45434755 | 1.26 |
ENSMUST00000233503.2
ENSMUST00000120005.10 ENSMUST00000211609.2 |
Lmtk3
|
lemur tyrosine kinase 3 |
| chr2_-_6327884 | 1.25 |
ENSMUST00000238876.2
|
1700014B07Rik
|
RIKEN cDNA 1700014B07 gene |
| chr8_+_111760521 | 1.25 |
ENSMUST00000034441.8
|
Aars
|
alanyl-tRNA synthetase |
| chr16_-_4867703 | 1.24 |
ENSMUST00000115844.3
ENSMUST00000023189.15 |
Glyr1
|
glyoxylate reductase 1 homolog (Arabidopsis) |
| chr4_+_154041687 | 1.24 |
ENSMUST00000105645.9
ENSMUST00000058393.9 ENSMUST00000141493.8 |
A430005L14Rik
|
RIKEN cDNA A430005L14 gene |
| chr7_+_44146029 | 1.23 |
ENSMUST00000205359.2
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr2_-_132089667 | 1.21 |
ENSMUST00000110163.8
ENSMUST00000180286.2 ENSMUST00000028816.9 |
Tmem230
|
transmembrane protein 230 |
| chr11_+_72686990 | 1.19 |
ENSMUST00000069395.7
ENSMUST00000172220.8 |
Zzef1
|
zinc finger, ZZ-type with EF hand domain 1 |
| chr6_-_134874778 | 1.18 |
ENSMUST00000165392.8
ENSMUST00000204880.3 ENSMUST00000203409.3 ENSMUST00000046255.14 ENSMUST00000111932.8 ENSMUST00000149375.8 ENSMUST00000116515.9 |
Gpr19
|
G protein-coupled receptor 19 |
| chr1_+_171910073 | 1.17 |
ENSMUST00000135192.8
|
Copa
|
coatomer protein complex subunit alpha |
| chr7_+_121666388 | 1.15 |
ENSMUST00000033158.6
|
Ubfd1
|
ubiquitin family domain containing 1 |
| chr8_+_46428551 | 1.14 |
ENSMUST00000034051.7
ENSMUST00000150943.2 |
Ufsp2
|
UFM1-specific peptidase 2 |
| chr11_+_52123016 | 1.13 |
ENSMUST00000109072.2
|
Skp1
|
S-phase kinase-associated protein 1 |
| chr1_+_185095232 | 1.12 |
ENSMUST00000046514.13
|
Eprs
|
glutamyl-prolyl-tRNA synthetase |
| chr1_-_183126875 | 1.11 |
ENSMUST00000195233.3
|
Mia3
|
melanoma inhibitory activity 3 |
| chr6_-_119825054 | 1.11 |
ENSMUST00000079582.5
|
Erc1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr4_-_150994260 | 1.10 |
ENSMUST00000134751.8
ENSMUST00000030805.14 |
Park7
|
Parkinson disease (autosomal recessive, early onset) 7 |
| chr9_+_108216466 | 1.09 |
ENSMUST00000193987.2
|
Gpx1
|
glutathione peroxidase 1 |
| chr17_-_26727437 | 1.09 |
ENSMUST00000236661.2
ENSMUST00000025025.7 |
Dusp1
|
dual specificity phosphatase 1 |
| chr9_-_50472605 | 1.06 |
ENSMUST00000034568.7
|
Tex12
|
testis expressed 12 |
| chr2_+_70491901 | 1.06 |
ENSMUST00000112201.8
ENSMUST00000028509.11 ENSMUST00000133432.8 ENSMUST00000112205.2 |
Gorasp2
|
golgi reassembly stacking protein 2 |
| chr5_+_107479023 | 1.06 |
ENSMUST00000031215.15
ENSMUST00000112677.10 |
Brdt
|
bromodomain, testis-specific |
| chr7_-_118132520 | 1.05 |
ENSMUST00000209146.2
ENSMUST00000098090.10 ENSMUST00000032887.4 |
Coq7
|
demethyl-Q 7 |
| chr18_+_65933586 | 1.04 |
ENSMUST00000025394.14
ENSMUST00000236847.2 ENSMUST00000153193.3 |
Sec11c
|
SEC11 homolog C, signal peptidase complex subunit |
| chr10_-_80895949 | 1.04 |
ENSMUST00000219401.2
ENSMUST00000220317.2 ENSMUST00000005067.6 ENSMUST00000218208.2 |
Sgta
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr16_-_13720949 | 1.03 |
ENSMUST00000023361.12
ENSMUST00000115802.2 |
Pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr2_+_71283620 | 1.03 |
ENSMUST00000037210.9
|
Metap1d
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr5_+_129970882 | 1.03 |
ENSMUST00000201855.2
ENSMUST00000073945.6 |
Vkorc1l1
|
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr16_+_36695479 | 1.02 |
ENSMUST00000023534.7
ENSMUST00000114812.9 ENSMUST00000134616.8 |
Golgb1
|
golgi autoantigen, golgin subfamily b, macrogolgin 1 |
| chr11_+_52122836 | 1.01 |
ENSMUST00000037324.12
ENSMUST00000166537.8 |
Skp1
|
S-phase kinase-associated protein 1 |
| chr6_+_29348068 | 1.00 |
ENSMUST00000173216.8
ENSMUST00000173694.5 ENSMUST00000172974.8 ENSMUST00000031779.17 ENSMUST00000090481.14 |
Calu
|
calumenin |
| chr4_+_109835224 | 1.00 |
ENSMUST00000061187.4
|
Dmrta2
|
doublesex and mab-3 related transcription factor like family A2 |
| chr16_-_13720915 | 0.99 |
ENSMUST00000115803.9
|
Pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr9_-_50472620 | 0.98 |
ENSMUST00000217236.2
|
Tex12
|
testis expressed 12 |
| chr3_-_154798982 | 0.98 |
ENSMUST00000066568.6
|
Fpgt
|
fucose-1-phosphate guanylyltransferase |
| chr10_-_121933276 | 0.97 |
ENSMUST00000140299.3
|
Rxylt1
|
ribitol xylosyltransferase 1 |
| chr8_-_34614187 | 0.96 |
ENSMUST00000033910.9
|
Leprotl1
|
leptin receptor overlapping transcript-like 1 |
| chr8_+_109441276 | 0.95 |
ENSMUST00000043896.10
|
Zfhx3
|
zinc finger homeobox 3 |
| chr7_+_16609227 | 0.95 |
ENSMUST00000108493.3
|
Dact3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr7_-_46445085 | 0.94 |
ENSMUST00000123725.2
|
Hps5
|
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr1_-_171910324 | 0.94 |
ENSMUST00000003550.11
|
Ncstn
|
nicastrin |
| chr11_+_54329014 | 0.93 |
ENSMUST00000046835.14
|
Fnip1
|
folliculin interacting protein 1 |
| chr1_-_93373145 | 0.92 |
ENSMUST00000186787.7
|
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr3_+_89153704 | 0.92 |
ENSMUST00000168900.3
|
Krtcap2
|
keratinocyte associated protein 2 |
| chr1_+_171910343 | 0.91 |
ENSMUST00000027833.12
|
Copa
|
coatomer protein complex subunit alpha |
| chrX_-_135642025 | 0.91 |
ENSMUST00000155207.8
ENSMUST00000080411.13 ENSMUST00000169418.8 |
Morf4l2
|
mortality factor 4 like 2 |
| chr17_-_84773544 | 0.90 |
ENSMUST00000047524.10
|
Thada
|
thyroid adenoma associated |
| chr1_+_130947580 | 0.89 |
ENSMUST00000016673.6
|
Il10
|
interleukin 10 |
| chr13_-_38712387 | 0.87 |
ENSMUST00000035988.16
|
Txndc5
|
thioredoxin domain containing 5 |
| chr2_-_90900628 | 0.86 |
ENSMUST00000111436.3
ENSMUST00000073575.12 |
Slc39a13
|
solute carrier family 39 (metal ion transporter), member 13 |
| chr17_-_28569721 | 0.84 |
ENSMUST00000156862.3
|
Tead3
|
TEA domain family member 3 |
| chr11_+_82671934 | 0.83 |
ENSMUST00000092849.12
ENSMUST00000021039.12 ENSMUST00000080461.12 ENSMUST00000173347.8 ENSMUST00000173727.8 ENSMUST00000173009.8 ENSMUST00000131537.9 ENSMUST00000173722.8 |
Lig3
|
ligase III, DNA, ATP-dependent |
| chr12_+_71356632 | 0.83 |
ENSMUST00000061273.12
ENSMUST00000150639.2 |
Dact1
|
dishevelled-binding antagonist of beta-catenin 1 |
| chr9_+_108216433 | 0.83 |
ENSMUST00000191997.2
|
Gpx1
|
glutathione peroxidase 1 |
| chr8_+_95328694 | 0.83 |
ENSMUST00000211983.2
|
Rspry1
|
ring finger and SPRY domain containing 1 |
| chr4_+_137321451 | 0.81 |
ENSMUST00000105840.8
ENSMUST00000105839.8 ENSMUST00000055131.13 ENSMUST00000105838.8 |
Usp48
|
ubiquitin specific peptidase 48 |
| chr1_+_119453849 | 0.81 |
ENSMUST00000183952.2
|
Tmem185b
|
transmembrane protein 185B |
| chr7_+_127511681 | 0.80 |
ENSMUST00000033070.9
|
Kat8
|
K(lysine) acetyltransferase 8 |
| chr18_-_84703738 | 0.79 |
ENSMUST00000025546.17
|
Cndp2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr16_-_13720863 | 0.79 |
ENSMUST00000115804.9
|
Pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chrX_+_106193060 | 0.79 |
ENSMUST00000125676.8
ENSMUST00000180182.2 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr17_+_34341766 | 0.78 |
ENSMUST00000042121.11
|
H2-DMa
|
histocompatibility 2, class II, locus DMa |
| chr11_+_69891398 | 0.77 |
ENSMUST00000019362.15
ENSMUST00000190940.2 |
Dvl2
|
dishevelled segment polarity protein 2 |
| chr10_+_77458197 | 0.77 |
ENSMUST00000172772.2
|
Ube2g2
|
ubiquitin-conjugating enzyme E2G 2 |
| chr2_-_132089543 | 0.77 |
ENSMUST00000110164.8
|
Tmem230
|
transmembrane protein 230 |
| chr15_-_99717956 | 0.76 |
ENSMUST00000109024.9
|
Lima1
|
LIM domain and actin binding 1 |
| chr3_+_30847024 | 0.74 |
ENSMUST00000029256.9
|
Sec62
|
SEC62 homolog (S. cerevisiae) |
| chr14_-_78774201 | 0.74 |
ENSMUST00000123853.9
|
Akap11
|
A kinase (PRKA) anchor protein 11 |
| chr7_+_45522551 | 0.74 |
ENSMUST00000211234.2
|
Kdelr1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr9_-_20888054 | 0.73 |
ENSMUST00000054197.7
|
S1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr3_+_154799223 | 0.72 |
ENSMUST00000194376.2
|
Lrriq3
|
leucine-rich repeats and IQ motif containing 3 |
| chr12_-_4088905 | 0.71 |
ENSMUST00000111178.2
|
Efr3b
|
EFR3 homolog B |
| chr2_+_23046381 | 0.70 |
ENSMUST00000028117.4
|
Yme1l1
|
YME1-like 1 (S. cerevisiae) |
| chr1_+_181952302 | 0.70 |
ENSMUST00000111018.2
ENSMUST00000027792.6 |
Srp9
|
signal recognition particle 9 |
| chr11_-_69871320 | 0.69 |
ENSMUST00000143175.2
|
Elp5
|
elongator acetyltransferase complex subunit 5 |
| chr5_+_120569764 | 0.68 |
ENSMUST00000031591.10
|
Lhx5
|
LIM homeobox protein 5 |
| chr15_+_34238174 | 0.68 |
ENSMUST00000022867.5
ENSMUST00000226627.2 |
Laptm4b
|
lysosomal-associated protein transmembrane 4B |
| chr14_+_79825131 | 0.68 |
ENSMUST00000054908.10
|
Sugt1
|
SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) |
| chr11_-_29975916 | 0.67 |
ENSMUST00000058902.6
|
Eml6
|
echinoderm microtubule associated protein like 6 |
| chr1_+_192855776 | 0.67 |
ENSMUST00000161235.3
ENSMUST00000160077.2 ENSMUST00000178744.2 ENSMUST00000192189.2 ENSMUST00000110831.4 ENSMUST00000191613.2 |
A130010J15Rik
|
RIKEN cDNA A130010J15 gene |
| chr2_+_154975419 | 0.67 |
ENSMUST00000029126.15
ENSMUST00000109685.8 |
Itch
|
itchy, E3 ubiquitin protein ligase |
| chr3_+_154799163 | 0.66 |
ENSMUST00000192383.6
|
Lrriq3
|
leucine-rich repeats and IQ motif containing 3 |
| chr18_+_67597929 | 0.66 |
ENSMUST00000025411.9
|
Prelid3a
|
PRELI domain containing 3A |
| chrX_-_7440480 | 0.66 |
ENSMUST00000115742.9
ENSMUST00000150787.8 |
Ppp1r3f
|
protein phosphatase 1, regulatory subunit 3F |
| chr18_-_31742946 | 0.64 |
ENSMUST00000060396.7
|
Slc25a46
|
solute carrier family 25, member 46 |
| chr8_+_95328558 | 0.64 |
ENSMUST00000060389.10
ENSMUST00000212729.2 ENSMUST00000121101.2 |
Rspry1
|
ring finger and SPRY domain containing 1 |
| chr18_-_35348049 | 0.63 |
ENSMUST00000091636.5
ENSMUST00000236680.2 |
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr17_-_34340918 | 0.62 |
ENSMUST00000151986.2
|
Brd2
|
bromodomain containing 2 |
| chr16_-_36695166 | 0.62 |
ENSMUST00000075946.12
|
Eaf2
|
ELL associated factor 2 |
| chrX_+_106193167 | 0.62 |
ENSMUST00000137107.2
ENSMUST00000067249.3 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr3_+_154799063 | 0.61 |
ENSMUST00000029833.13
|
Lrriq3
|
leucine-rich repeats and IQ motif containing 3 |
| chr13_-_100969878 | 0.61 |
ENSMUST00000067246.6
|
Slc30a5
|
solute carrier family 30 (zinc transporter), member 5 |
| chr13_-_54836077 | 0.60 |
ENSMUST00000150626.2
ENSMUST00000134177.8 |
Rnf44
|
ring finger protein 44 |
| chr10_+_57508203 | 0.60 |
ENSMUST00000177438.8
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr9_-_103243039 | 0.59 |
ENSMUST00000035484.11
|
Cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
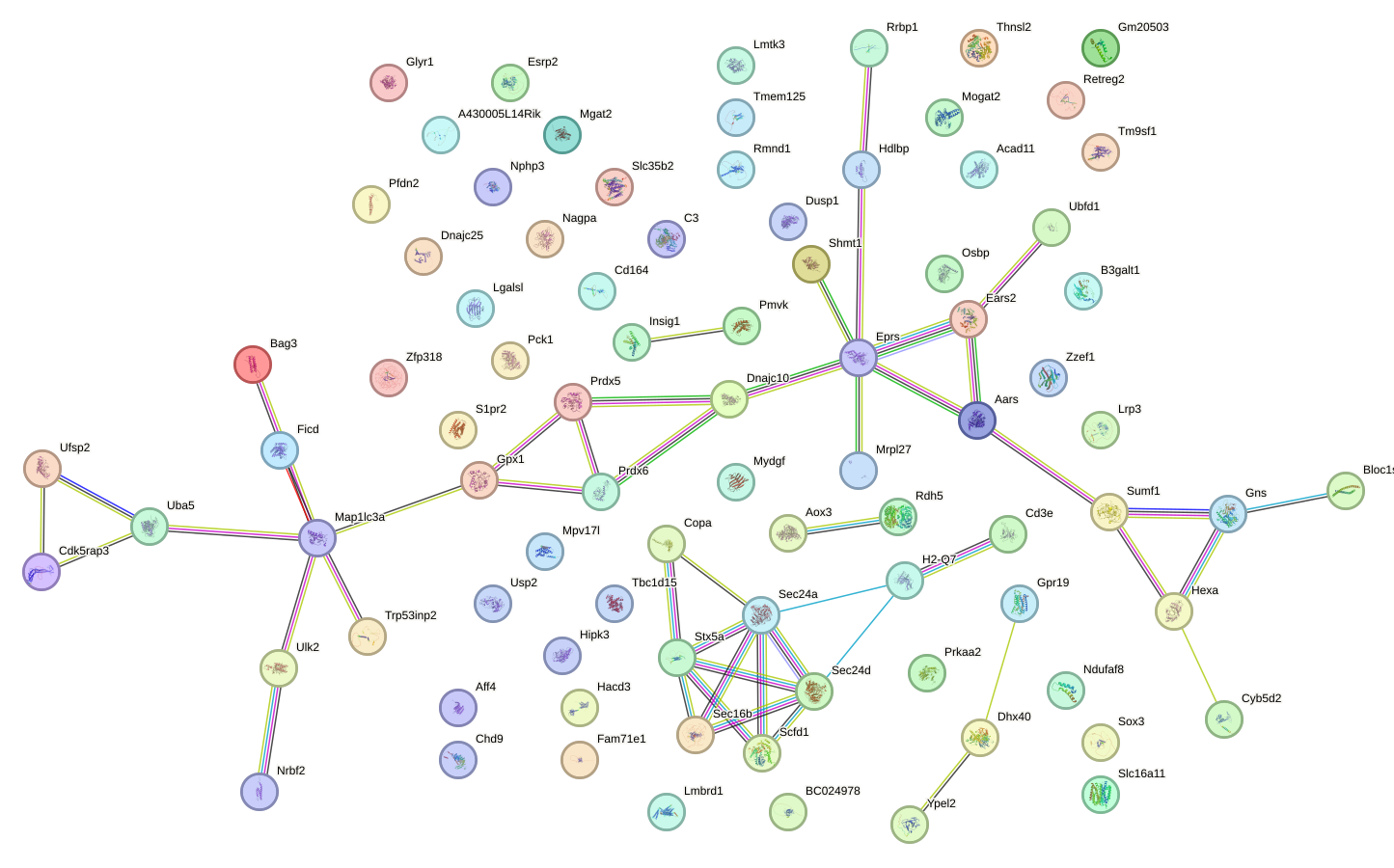
Network of associatons between targets according to the STRING database.
First level regulatory network of Creb3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 8.2 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 1.6 | 6.3 | GO:0006114 | glycerol biosynthetic process(GO:0006114) positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 1.5 | 6.1 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 1.4 | 11.6 | GO:0071569 | protein ufmylation(GO:0071569) |
| 1.0 | 4.2 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.9 | 17.0 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.9 | 3.7 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.8 | 2.5 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.8 | 3.8 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.7 | 7.7 | GO:1903298 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.7 | 4.8 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.7 | 3.9 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.6 | 3.2 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.6 | 7.7 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.6 | 2.2 | GO:0010269 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) response to selenium ion(GO:0010269) |
| 0.5 | 2.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.4 | 4.3 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.4 | 1.2 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.4 | 4.2 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.4 | 2.6 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.4 | 3.6 | GO:0075044 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.3 | 2.0 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.3 | 3.4 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.3 | 0.9 | GO:0060300 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) regulation of cytokine activity(GO:0060300) |
| 0.3 | 1.7 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.3 | 1.4 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.3 | 0.8 | GO:0090298 | negative regulation of mitochondrial DNA replication(GO:0090298) |
| 0.3 | 1.1 | GO:1902958 | enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) glycolate biosynthetic process(GO:0046295) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.3 | 1.6 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.3 | 1.1 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.3 | 0.8 | GO:0002501 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.3 | 1.0 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.3 | 1.0 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.3 | 2.6 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.2 | 4.3 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.2 | 0.7 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.2 | 0.6 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.2 | 1.8 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.2 | 0.6 | GO:1902868 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.2 | 0.8 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.2 | 1.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.2 | 1.7 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 4.7 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.2 | 4.4 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.2 | 1.4 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.2 | 8.2 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 7.1 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 1.0 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 0.7 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.6 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.1 | 0.9 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.1 | 1.4 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 0.5 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.1 | 0.4 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 1.6 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.7 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.1 | 1.8 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.7 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.1 | 4.6 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.1 | 0.8 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 0.9 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 0.8 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.1 | 0.3 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.1 | 0.8 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 0.4 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 6.3 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 1.7 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) |
| 0.1 | 0.7 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 2.4 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 1.4 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.2 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 1.0 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 1.9 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 1.5 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 0.4 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.1 | 1.0 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.3 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 1.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.6 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.9 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.7 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 | 2.4 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 1.1 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 1.0 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.9 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 1.3 | GO:0007530 | sex determination(GO:0007530) |
| 0.0 | 0.9 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.0 | 0.8 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 1.7 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0021763 | subthalamic nucleus development(GO:0021763) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.5 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 1.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.2 | GO:0098970 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.0 | 0.9 | GO:0044819 | mitotic G1 DNA damage checkpoint(GO:0031571) mitotic G1/S transition checkpoint(GO:0044819) |
| 0.0 | 6.9 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 2.2 | GO:1902652 | cholesterol metabolic process(GO:0008203) secondary alcohol metabolic process(GO:1902652) |
| 0.0 | 1.1 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 0.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.4 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 2.3 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.4 | GO:0061088 | cobalt ion transport(GO:0006824) sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 1.1 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.6 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 1.1 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 1.8 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 | 0.4 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.2 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 2.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0016093 | polyprenol metabolic process(GO:0016093) |
| 0.0 | 1.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 1.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.3 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 1.4 | GO:0030203 | glycosaminoglycan metabolic process(GO:0030203) |
| 0.0 | 0.7 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 2.4 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 2.0 | GO:0048489 | synaptic vesicle transport(GO:0048489) establishment of synaptic vesicle localization(GO:0097480) |
| 0.0 | 2.1 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.6 | GO:0051452 | intracellular pH reduction(GO:0051452) |
| 0.0 | 1.5 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.0 | GO:1904393 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) |
| 0.0 | 0.4 | GO:0032456 | endocytic recycling(GO:0032456) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.6 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.6 | 3.2 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.6 | 9.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.6 | 10.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.5 | 7.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.3 | 2.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.3 | 2.4 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.3 | 2.4 | GO:0097413 | Lewy body(GO:0097413) |
| 0.3 | 4.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 0.9 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.2 | 3.4 | GO:0044754 | autolysosome(GO:0044754) |
| 0.2 | 3.6 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 0.6 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.2 | 6.1 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 4.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.2 | 0.5 | GO:0055087 | Ski complex(GO:0055087) |
| 0.2 | 2.0 | GO:0000801 | central element(GO:0000801) |
| 0.2 | 1.9 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 4.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 2.0 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 1.7 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.6 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 0.7 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 1.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.8 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 1.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.7 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 0.7 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 0.6 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 3.7 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 4.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 1.4 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 1.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.9 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 1.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.2 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 0.1 | 1.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.8 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.9 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 2.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 10.9 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.5 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 1.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.2 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 1.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 2.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.6 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 5.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 2.4 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 5.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.6 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 4.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 7.9 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 3.0 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.0 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 6.2 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 1.9 | GO:0030018 | Z disc(GO:0030018) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.3 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 1.6 | 8.2 | GO:0030151 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) molybdenum ion binding(GO:0030151) |
| 1.6 | 6.3 | GO:0070905 | serine binding(GO:0070905) |
| 1.6 | 4.7 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 1.5 | 6.1 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 1.5 | 4.4 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.6 | 1.8 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.4 | 1.7 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.4 | 7.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.4 | 1.2 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.4 | 1.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.4 | 9.9 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.4 | 1.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.3 | 1.0 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.3 | 3.7 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.3 | 3.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.3 | 7.7 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.3 | 1.4 | GO:0102345 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.3 | 1.4 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.3 | 1.1 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.2 | 4.3 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.2 | 1.0 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 1.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 0.6 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.2 | 0.7 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.2 | 1.9 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 2.6 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.2 | 1.0 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 0.9 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.7 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 1.8 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.7 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.4 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 3.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 5.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.5 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.1 | 0.4 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 1.7 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.1 | 1.4 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.1 | 0.4 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.1 | 1.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 0.8 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 3.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 1.2 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 2.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 2.6 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 0.8 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 0.4 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.1 | 0.6 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 2.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 2.7 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 0.6 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 1.1 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 2.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 1.7 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.9 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.7 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.5 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 5.9 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.3 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.8 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 5.1 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.8 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 1.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 1.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 2.0 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 1.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.4 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 4.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.2 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.6 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.0 | 0.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.1 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 1.3 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.9 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 4.0 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 0.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 1.1 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 0.1 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 1.5 | GO:0008565 | protein transporter activity(GO:0008565) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 4.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 2.9 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 2.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 6.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.6 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 4.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.8 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 2.0 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 2.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 2.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.7 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.3 | 4.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.3 | 6.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.3 | 4.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.3 | 2.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 2.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 2.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 1.9 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.2 | 2.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 4.4 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.1 | 3.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 2.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 2.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 3.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 2.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 0.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 0.8 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 3.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.6 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.9 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 1.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 2.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 3.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.3 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 3.4 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 2.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.4 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |