Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
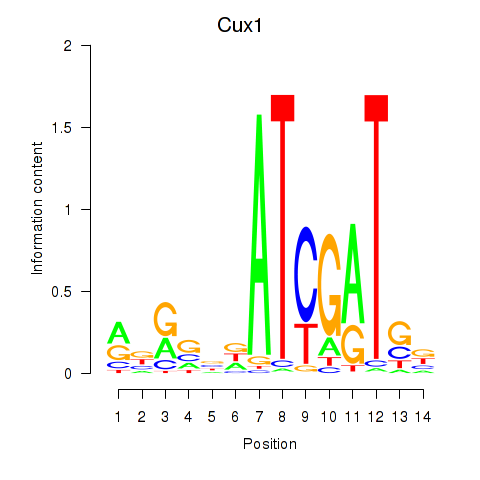
Results for Cux1
Z-value: 1.34
Transcription factors associated with Cux1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cux1
|
ENSMUSG00000029705.18 | cut-like homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cux1 | mm39_v1_chr5_-_136596299_136596390 | -0.52 | 1.1e-03 | Click! |
Activity profile of Cux1 motif
Sorted Z-values of Cux1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_39275518 | 12.99 |
ENSMUST00000003137.15
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr19_+_38995463 | 11.71 |
ENSMUST00000025966.5
|
Cyp2c55
|
cytochrome P450, family 2, subfamily c, polypeptide 55 |
| chr7_-_99345016 | 10.89 |
ENSMUST00000107086.9
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr19_-_44396092 | 10.74 |
ENSMUST00000041331.4
|
Scd1
|
stearoyl-Coenzyme A desaturase 1 |
| chr4_-_62069046 | 8.51 |
ENSMUST00000077719.4
|
Mup21
|
major urinary protein 21 |
| chr13_-_4573312 | 8.15 |
ENSMUST00000221564.2
ENSMUST00000078239.5 ENSMUST00000080361.13 |
Akr1c20
|
aldo-keto reductase family 1, member C20 |
| chr8_-_94006345 | 7.85 |
ENSMUST00000034178.9
|
Ces1f
|
carboxylesterase 1F |
| chr7_+_26819334 | 7.67 |
ENSMUST00000003100.10
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2 |
| chr7_-_99344779 | 6.70 |
ENSMUST00000137914.2
ENSMUST00000207090.2 ENSMUST00000208225.2 |
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr19_-_40062174 | 6.40 |
ENSMUST00000048959.5
|
Cyp2c54
|
cytochrome P450, family 2, subfamily c, polypeptide 54 |
| chr19_-_44396269 | 6.36 |
ENSMUST00000235741.2
|
Scd1
|
stearoyl-Coenzyme A desaturase 1 |
| chr4_+_115156243 | 6.34 |
ENSMUST00000084343.4
|
Cyp4a12a
|
cytochrome P450, family 4, subfamily a, polypeptide 12a |
| chr8_-_93956143 | 6.06 |
ENSMUST00000176282.2
ENSMUST00000034173.14 |
Ces1e
|
carboxylesterase 1E |
| chr17_+_24955613 | 5.88 |
ENSMUST00000115262.9
|
Msrb1
|
methionine sulfoxide reductase B1 |
| chr11_+_78389913 | 5.77 |
ENSMUST00000017488.5
|
Vtn
|
vitronectin |
| chr19_+_39980868 | 5.69 |
ENSMUST00000049178.3
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr7_-_99344832 | 5.66 |
ENSMUST00000145381.8
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr16_+_22739028 | 5.50 |
ENSMUST00000232097.2
|
Fetub
|
fetuin beta |
| chr9_-_118986123 | 5.36 |
ENSMUST00000010795.5
|
Acaa1b
|
acetyl-Coenzyme A acyltransferase 1B |
| chr5_-_89605622 | 5.27 |
ENSMUST00000049209.13
|
Gc
|
vitamin D binding protein |
| chr8_+_110717062 | 5.25 |
ENSMUST00000001720.14
ENSMUST00000143741.2 |
Tat
|
tyrosine aminotransferase |
| chr9_+_46179899 | 5.18 |
ENSMUST00000121598.8
|
Apoa5
|
apolipoprotein A-V |
| chr7_-_19432308 | 5.16 |
ENSMUST00000173739.8
|
Apoe
|
apolipoprotein E |
| chr17_-_35077089 | 4.76 |
ENSMUST00000153400.8
|
Cfb
|
complement factor B |
| chr11_-_96807192 | 4.59 |
ENSMUST00000144731.8
ENSMUST00000127048.8 |
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr1_+_58152295 | 4.56 |
ENSMUST00000040999.14
ENSMUST00000162011.3 |
Aox3
|
aldehyde oxidase 3 |
| chr17_+_24955647 | 4.49 |
ENSMUST00000101800.7
|
Msrb1
|
methionine sulfoxide reductase B1 |
| chr11_-_96807273 | 4.42 |
ENSMUST00000103152.11
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr6_-_128503666 | 4.42 |
ENSMUST00000143664.2
ENSMUST00000112132.8 |
Pzp
|
PZP, alpha-2-macroglobulin like |
| chr1_+_131725119 | 3.91 |
ENSMUST00000112393.9
ENSMUST00000048660.12 |
Pm20d1
|
peptidase M20 domain containing 1 |
| chr16_+_22739191 | 3.86 |
ENSMUST00000116625.10
|
Fetub
|
fetuin beta |
| chr6_+_121323577 | 3.85 |
ENSMUST00000032200.16
|
Slc6a12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 |
| chrM_+_10167 | 3.79 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr10_-_126956991 | 3.65 |
ENSMUST00000080975.6
ENSMUST00000164259.9 |
Os9
|
amplified in osteosarcoma |
| chr11_-_70910058 | 3.64 |
ENSMUST00000108523.10
ENSMUST00000143850.8 |
Derl2
|
Der1-like domain family, member 2 |
| chr11_-_96807233 | 3.57 |
ENSMUST00000130774.2
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr1_+_180878797 | 3.48 |
ENSMUST00000036819.7
|
9130409I23Rik
|
RIKEN cDNA 9130409I23 gene |
| chr7_-_99340830 | 3.45 |
ENSMUST00000208713.2
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr9_-_103182246 | 3.33 |
ENSMUST00000142540.2
|
1300017J02Rik
|
RIKEN cDNA 1300017J02 gene |
| chr4_+_104623505 | 3.29 |
ENSMUST00000031663.10
ENSMUST00000065072.7 |
C8b
|
complement component 8, beta polypeptide |
| chr13_+_4109566 | 3.27 |
ENSMUST00000041768.7
|
Akr1c14
|
aldo-keto reductase family 1, member C14 |
| chr8_-_110305672 | 3.26 |
ENSMUST00000074898.8
|
Hp
|
haptoglobin |
| chr7_+_44033520 | 3.26 |
ENSMUST00000118962.8
ENSMUST00000118831.8 |
Syt3
|
synaptotagmin III |
| chr3_-_157630690 | 3.25 |
ENSMUST00000118539.2
|
Cth
|
cystathionase (cystathionine gamma-lyase) |
| chr12_-_103704417 | 3.24 |
ENSMUST00000095450.11
ENSMUST00000187220.2 |
Serpina1b
|
serine (or cysteine) preptidase inhibitor, clade A, member 1B |
| chr5_-_147831610 | 3.21 |
ENSMUST00000118527.8
ENSMUST00000031655.4 ENSMUST00000138244.2 |
Slc46a3
|
solute carrier family 46, member 3 |
| chr5_-_87572060 | 3.11 |
ENSMUST00000072818.6
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr19_-_4889314 | 3.07 |
ENSMUST00000235245.2
ENSMUST00000037246.7 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr12_-_103829810 | 3.05 |
ENSMUST00000085056.8
ENSMUST00000072876.12 ENSMUST00000124717.2 |
Serpina1a
|
serine (or cysteine) peptidase inhibitor, clade A, member 1A |
| chr3_-_63872189 | 3.05 |
ENSMUST00000029402.15
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr17_+_84990541 | 3.02 |
ENSMUST00000045714.15
ENSMUST00000171915.2 |
Abcg8
|
ATP binding cassette subfamily G member 8 |
| chr9_-_26910833 | 3.01 |
ENSMUST00000060513.8
ENSMUST00000120367.8 |
Acad8
|
acyl-Coenzyme A dehydrogenase family, member 8 |
| chr1_+_87998487 | 2.97 |
ENSMUST00000073772.5
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr15_-_100583044 | 2.95 |
ENSMUST00000230312.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr19_-_4889284 | 2.94 |
ENSMUST00000236451.2
ENSMUST00000236178.2 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr2_-_23939401 | 2.90 |
ENSMUST00000051416.12
|
Hnmt
|
histamine N-methyltransferase |
| chr3_+_94280101 | 2.73 |
ENSMUST00000029795.10
|
Rorc
|
RAR-related orphan receptor gamma |
| chr5_+_87148697 | 2.72 |
ENSMUST00000031186.9
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr11_-_106471420 | 2.69 |
ENSMUST00000153870.2
|
Tex2
|
testis expressed gene 2 |
| chr18_-_60860594 | 2.68 |
ENSMUST00000235795.2
|
Ndst1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr11_+_69945157 | 2.64 |
ENSMUST00000108585.9
ENSMUST00000018699.13 |
Asgr1
|
asialoglycoprotein receptor 1 |
| chrX_-_8059597 | 2.64 |
ENSMUST00000143223.2
ENSMUST00000033509.15 |
Ebp
|
phenylalkylamine Ca2+ antagonist (emopamil) binding protein |
| chr11_+_51654511 | 2.55 |
ENSMUST00000020653.6
|
Sar1b
|
secretion associated Ras related GTPase 1B |
| chr1_-_179373826 | 2.55 |
ENSMUST00000027769.6
|
Tfb2m
|
transcription factor B2, mitochondrial |
| chrX_+_7588505 | 2.54 |
ENSMUST00000207675.2
ENSMUST00000116633.9 ENSMUST00000208996.2 ENSMUST00000144148.4 ENSMUST00000125991.9 ENSMUST00000148624.8 |
Wdr45
|
WD repeat domain 45 |
| chr7_+_142606476 | 2.50 |
ENSMUST00000037941.10
|
Cd81
|
CD81 antigen |
| chr4_-_63072367 | 2.45 |
ENSMUST00000030041.5
|
Ambp
|
alpha 1 microglobulin/bikunin precursor |
| chr4_+_62278932 | 2.44 |
ENSMUST00000084526.12
|
Slc31a1
|
solute carrier family 31, member 1 |
| chr7_+_44034225 | 2.42 |
ENSMUST00000120262.2
|
Syt3
|
synaptotagmin III |
| chr6_-_124718316 | 2.41 |
ENSMUST00000004389.6
|
Grcc10
|
gene rich cluster, C10 gene |
| chr12_-_103923145 | 2.39 |
ENSMUST00000085054.5
|
Serpina1e
|
serine (or cysteine) peptidase inhibitor, clade A, member 1E |
| chr1_+_16758629 | 2.38 |
ENSMUST00000026881.11
|
Ly96
|
lymphocyte antigen 96 |
| chrX_+_7588453 | 2.37 |
ENSMUST00000043045.10
ENSMUST00000207386.2 ENSMUST00000116634.9 ENSMUST00000208072.2 ENSMUST00000207589.2 ENSMUST00000208618.2 ENSMUST00000208443.2 ENSMUST00000207541.2 ENSMUST00000208528.2 ENSMUST00000115689.10 ENSMUST00000131077.9 ENSMUST00000115688.8 ENSMUST00000208156.2 |
Wdr45
Gm45208
|
WD repeat domain 45 predicted gene 45208 |
| chr9_+_103940575 | 2.32 |
ENSMUST00000120854.8
|
Acad11
|
acyl-Coenzyme A dehydrogenase family, member 11 |
| chr1_+_88128323 | 2.30 |
ENSMUST00000049289.9
|
Ugt1a2
|
UDP glucuronosyltransferase 1 family, polypeptide A2 |
| chrX_+_100420873 | 2.27 |
ENSMUST00000052130.14
|
Gjb1
|
gap junction protein, beta 1 |
| chr2_-_84605732 | 2.25 |
ENSMUST00000023994.10
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr11_+_73090270 | 2.23 |
ENSMUST00000006105.7
|
Shpk
|
sedoheptulokinase |
| chr2_-_84605764 | 2.22 |
ENSMUST00000111641.2
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr1_+_107289659 | 2.17 |
ENSMUST00000027566.3
|
Serpinb11
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 11 |
| chr7_+_130633776 | 2.17 |
ENSMUST00000084509.7
ENSMUST00000213064.3 ENSMUST00000208311.4 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr10_+_80165787 | 2.15 |
ENSMUST00000105358.8
ENSMUST00000105357.2 ENSMUST00000105354.8 ENSMUST00000105355.8 |
Reep6
|
receptor accessory protein 6 |
| chr9_-_50657800 | 2.14 |
ENSMUST00000239417.2
ENSMUST00000034564.4 |
2310030G06Rik
|
RIKEN cDNA 2310030G06 gene |
| chrX_+_100427331 | 2.13 |
ENSMUST00000119190.2
|
Gjb1
|
gap junction protein, beta 1 |
| chr18_+_50261268 | 2.11 |
ENSMUST00000025385.7
|
Hsd17b4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr5_+_3646066 | 2.08 |
ENSMUST00000006061.13
ENSMUST00000121291.8 ENSMUST00000142516.2 |
Pex1
|
peroxisomal biogenesis factor 1 |
| chr9_-_100916946 | 2.07 |
ENSMUST00000035116.12
|
Pccb
|
propionyl Coenzyme A carboxylase, beta polypeptide |
| chr6_+_88701810 | 2.02 |
ENSMUST00000089449.5
|
Mgll
|
monoglyceride lipase |
| chr18_-_56695333 | 2.02 |
ENSMUST00000066208.13
ENSMUST00000172734.8 |
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr3_-_89186940 | 2.01 |
ENSMUST00000118860.2
ENSMUST00000029566.9 |
Efna1
|
ephrin A1 |
| chrX_+_141011173 | 2.01 |
ENSMUST00000112914.8
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr1_+_16758731 | 2.00 |
ENSMUST00000190366.2
|
Ly96
|
lymphocyte antigen 96 |
| chr5_-_87686048 | 2.00 |
ENSMUST00000031199.11
|
Sult1b1
|
sulfotransferase family 1B, member 1 |
| chr9_-_100916910 | 2.00 |
ENSMUST00000142676.8
ENSMUST00000149322.8 ENSMUST00000189498.2 |
Pccb
|
propionyl Coenzyme A carboxylase, beta polypeptide |
| chr2_-_165997551 | 1.99 |
ENSMUST00000109249.9
ENSMUST00000146497.9 |
Sulf2
|
sulfatase 2 |
| chr2_-_165997179 | 1.99 |
ENSMUST00000088086.4
|
Sulf2
|
sulfatase 2 |
| chr4_+_43493344 | 1.97 |
ENSMUST00000030181.12
ENSMUST00000107922.3 |
Ccdc107
|
coiled-coil domain containing 107 |
| chr7_+_44240310 | 1.95 |
ENSMUST00000107906.6
|
Kcnc3
|
potassium voltage gated channel, Shaw-related subfamily, member 3 |
| chr16_+_70110837 | 1.93 |
ENSMUST00000163832.8
|
Gbe1
|
glucan (1,4-alpha-), branching enzyme 1 |
| chr13_+_93810911 | 1.93 |
ENSMUST00000048001.8
|
Dmgdh
|
dimethylglycine dehydrogenase precursor |
| chr15_+_88703786 | 1.92 |
ENSMUST00000024042.5
|
Creld2
|
cysteine-rich with EGF-like domains 2 |
| chr14_-_118370144 | 1.90 |
ENSMUST00000022727.10
ENSMUST00000228543.2 |
Tgds
|
TDP-glucose 4,6-dehydratase |
| chrX_+_150784841 | 1.88 |
ENSMUST00000026289.10
ENSMUST00000112617.4 |
Hsd17b10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr1_-_162726234 | 1.88 |
ENSMUST00000111510.8
ENSMUST00000045902.13 |
Fmo2
|
flavin containing monooxygenase 2 |
| chr11_-_106378622 | 1.87 |
ENSMUST00000001059.9
ENSMUST00000106799.2 ENSMUST00000106800.2 |
Ern1
|
endoplasmic reticulum (ER) to nucleus signalling 1 |
| chr7_+_29883611 | 1.83 |
ENSMUST00000208441.2
|
Cox7a1
|
cytochrome c oxidase subunit 7A1 |
| chrM_+_9870 | 1.82 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr6_+_78402956 | 1.81 |
ENSMUST00000079926.6
|
Reg1
|
regenerating islet-derived 1 |
| chr2_+_151947444 | 1.80 |
ENSMUST00000041500.8
|
Srxn1
|
sulfiredoxin 1 homolog (S. cerevisiae) |
| chr10_+_80165961 | 1.78 |
ENSMUST00000186864.7
ENSMUST00000040081.7 |
Reep6
|
receptor accessory protein 6 |
| chr17_+_31652029 | 1.78 |
ENSMUST00000136384.9
|
Pde9a
|
phosphodiesterase 9A |
| chr14_+_69409251 | 1.76 |
ENSMUST00000062437.10
|
Nkx2-6
|
NK2 homeobox 6 |
| chr19_+_38469504 | 1.74 |
ENSMUST00000182481.8
|
Plce1
|
phospholipase C, epsilon 1 |
| chr16_+_22676589 | 1.73 |
ENSMUST00000004574.14
ENSMUST00000178320.2 ENSMUST00000166487.10 |
Dnajb11
|
DnaJ heat shock protein family (Hsp40) member B11 |
| chr13_-_34314328 | 1.72 |
ENSMUST00000075774.5
|
Tubb2b
|
tubulin, beta 2B class IIB |
| chr3_-_89009214 | 1.72 |
ENSMUST00000081848.13
|
Fdps
|
farnesyl diphosphate synthetase |
| chr3_-_63872079 | 1.71 |
ENSMUST00000161659.8
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr14_+_118374511 | 1.70 |
ENSMUST00000022728.4
|
Gpr180
|
G protein-coupled receptor 180 |
| chr3_-_88204286 | 1.70 |
ENSMUST00000107556.10
|
Tsacc
|
TSSK6 activating co-chaperone |
| chr11_-_60243695 | 1.69 |
ENSMUST00000095254.12
ENSMUST00000102683.11 ENSMUST00000093048.13 ENSMUST00000093046.13 ENSMUST00000064019.15 ENSMUST00000102682.5 |
Tom1l2
|
target of myb1-like 2 (chicken) |
| chr3_-_88204145 | 1.68 |
ENSMUST00000010682.4
|
Tsacc
|
TSSK6 activating co-chaperone |
| chr11_-_82655132 | 1.68 |
ENSMUST00000021040.10
ENSMUST00000100722.5 |
Cct6b
|
chaperonin containing Tcp1, subunit 6b (zeta) |
| chr3_-_89009153 | 1.64 |
ENSMUST00000199668.3
ENSMUST00000196709.5 |
Fdps
|
farnesyl diphosphate synthetase |
| chr3_-_113198765 | 1.61 |
ENSMUST00000179568.3
|
Amy2a4
|
amylase 2a4 |
| chr16_-_90731394 | 1.60 |
ENSMUST00000142340.2
|
Cfap298
|
cilia and flagella associate protien 298 |
| chr8_-_78337226 | 1.60 |
ENSMUST00000034030.15
|
Tmem184c
|
transmembrane protein 184C |
| chr7_-_140462187 | 1.60 |
ENSMUST00000211179.2
|
Sirt3
|
sirtuin 3 |
| chr12_-_112905721 | 1.59 |
ENSMUST00000221497.2
ENSMUST00000002881.5 ENSMUST00000221397.2 |
Nudt14
|
nudix (nucleoside diphosphate linked moiety X)-type motif 14 |
| chr13_-_67081138 | 1.59 |
ENSMUST00000021991.11
|
Mterf3
|
mitochondrial transcription termination factor 3 |
| chr10_-_24587884 | 1.58 |
ENSMUST00000135846.2
|
Enpp1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chrX_+_141010919 | 1.57 |
ENSMUST00000042329.12
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr19_+_46587523 | 1.56 |
ENSMUST00000138302.9
ENSMUST00000099376.11 |
Wbp1l
|
WW domain binding protein 1 like |
| chr7_-_140462221 | 1.56 |
ENSMUST00000026559.14
|
Sirt3
|
sirtuin 3 |
| chr11_-_50216426 | 1.55 |
ENSMUST00000179865.8
ENSMUST00000020637.9 |
Canx
|
calnexin |
| chr13_-_67080968 | 1.53 |
ENSMUST00000172597.8
ENSMUST00000173773.2 |
Mterf3
|
mitochondrial transcription termination factor 3 |
| chr7_+_16186704 | 1.51 |
ENSMUST00000019302.10
|
Tmem160
|
transmembrane protein 160 |
| chr8_+_11547506 | 1.50 |
ENSMUST00000177955.8
ENSMUST00000033901.11 ENSMUST00000178721.2 |
Naxd
|
NAD(P)HX dehydratase |
| chr3_-_116388334 | 1.50 |
ENSMUST00000197190.5
ENSMUST00000198454.2 |
Trmt13
|
tRNA methyltransferase 13 |
| chr16_+_29398149 | 1.47 |
ENSMUST00000160597.8
|
Opa1
|
OPA1, mitochondrial dynamin like GTPase |
| chr14_-_25769457 | 1.47 |
ENSMUST00000069180.8
|
Zcchc24
|
zinc finger, CCHC domain containing 24 |
| chr16_+_29398165 | 1.46 |
ENSMUST00000161186.8
ENSMUST00000038867.13 |
Opa1
|
OPA1, mitochondrial dynamin like GTPase |
| chr15_-_11399666 | 1.45 |
ENSMUST00000022849.7
|
Tars
|
threonyl-tRNA synthetase |
| chr7_+_4240697 | 1.45 |
ENSMUST00000117550.2
|
Lilra5
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 |
| chr11_-_59937302 | 1.44 |
ENSMUST00000000310.14
ENSMUST00000102693.9 ENSMUST00000148512.2 |
Pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr3_+_63202687 | 1.44 |
ENSMUST00000194836.6
ENSMUST00000191633.6 |
Mme
|
membrane metallo endopeptidase |
| chr11_-_53313950 | 1.44 |
ENSMUST00000036045.6
|
Leap2
|
liver-expressed antimicrobial peptide 2 |
| chr3_+_20011201 | 1.43 |
ENSMUST00000091309.12
ENSMUST00000108329.8 ENSMUST00000003714.13 |
Cp
|
ceruloplasmin |
| chrM_+_3906 | 1.43 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr6_-_24527545 | 1.41 |
ENSMUST00000118558.5
|
Ndufa5
|
NADH:ubiquinone oxidoreductase subunit A5 |
| chr16_+_96162854 | 1.41 |
ENSMUST00000113794.8
|
Igsf5
|
immunoglobulin superfamily, member 5 |
| chr8_-_78337297 | 1.41 |
ENSMUST00000141202.2
ENSMUST00000152168.8 |
Tmem184c
|
transmembrane protein 184C |
| chr10_-_71180763 | 1.41 |
ENSMUST00000045887.9
|
Cisd1
|
CDGSH iron sulfur domain 1 |
| chr15_-_89263790 | 1.40 |
ENSMUST00000238996.2
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr2_-_165996716 | 1.40 |
ENSMUST00000139266.2
|
Sulf2
|
sulfatase 2 |
| chr11_-_95966407 | 1.40 |
ENSMUST00000107686.8
ENSMUST00000107684.2 |
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
| chr6_-_88021999 | 1.40 |
ENSMUST00000113598.8
|
Rab7
|
RAB7, member RAS oncogene family |
| chr7_-_137012444 | 1.39 |
ENSMUST00000120340.2
ENSMUST00000117404.8 ENSMUST00000068996.13 |
9430038I01Rik
|
RIKEN cDNA 9430038I01 gene |
| chr2_-_130506484 | 1.39 |
ENSMUST00000089559.11
|
Ddrgk1
|
DDRGK domain containing 1 |
| chr16_+_34842764 | 1.38 |
ENSMUST00000061156.10
|
Hacd2
|
3-hydroxyacyl-CoA dehydratase 2 |
| chr1_+_172525613 | 1.38 |
ENSMUST00000038495.5
|
Crp
|
C-reactive protein, pentraxin-related |
| chr3_-_113166153 | 1.38 |
ENSMUST00000098673.5
|
Amy2a5
|
amylase 2a5 |
| chr14_-_9184969 | 1.37 |
ENSMUST00000070323.12
|
Synpr
|
synaptoporin |
| chr16_-_90731611 | 1.36 |
ENSMUST00000149833.8
|
Cfap298
|
cilia and flagella associate protien 298 |
| chr11_-_120604551 | 1.35 |
ENSMUST00000106154.8
ENSMUST00000106155.4 ENSMUST00000055424.13 ENSMUST00000026137.8 |
Cenpx
|
centromere protein X |
| chr9_-_56835633 | 1.34 |
ENSMUST00000050916.7
|
Snx33
|
sorting nexin 33 |
| chr1_+_171173237 | 1.33 |
ENSMUST00000135941.8
|
Pfdn2
|
prefoldin 2 |
| chr12_+_103498542 | 1.33 |
ENSMUST00000021631.12
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr7_-_27373939 | 1.33 |
ENSMUST00000138243.2
|
Map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr6_+_30639217 | 1.32 |
ENSMUST00000031806.10
|
Cpa1
|
carboxypeptidase A1, pancreatic |
| chr5_-_91550853 | 1.31 |
ENSMUST00000121044.6
|
Btc
|
betacellulin, epidermal growth factor family member |
| chr17_-_35265514 | 1.31 |
ENSMUST00000007250.14
|
Msh5
|
mutS homolog 5 |
| chr3_+_20011251 | 1.30 |
ENSMUST00000108328.8
|
Cp
|
ceruloplasmin |
| chr3_-_58433313 | 1.30 |
ENSMUST00000029385.9
|
Serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr8_-_3767547 | 1.29 |
ENSMUST00000058040.7
|
Clec4g
|
C-type lectin domain family 4, member g |
| chr19_+_45064539 | 1.28 |
ENSMUST00000026234.5
|
Kazald1
|
Kazal-type serine peptidase inhibitor domain 1 |
| chrX_-_100838004 | 1.28 |
ENSMUST00000147742.9
|
Gm4779
|
predicted gene 4779 |
| chr10_-_17898838 | 1.27 |
ENSMUST00000220433.2
|
Abracl
|
ABRA C-terminal like |
| chr10_+_79505203 | 1.25 |
ENSMUST00000020552.8
ENSMUST00000239401.2 |
Tpgs1
|
tubulin polyglutamylase complex subunit 1 |
| chr1_-_183078488 | 1.24 |
ENSMUST00000057062.12
|
Brox
|
BRO1 domain and CAAX motif containing |
| chr9_-_108911438 | 1.23 |
ENSMUST00000192801.2
|
Tma7
|
translational machinery associated 7 |
| chr13_+_67080864 | 1.23 |
ENSMUST00000021990.4
|
Ptdss1
|
phosphatidylserine synthase 1 |
| chr17_-_33979280 | 1.23 |
ENSMUST00000173860.8
|
Rab11b
|
RAB11B, member RAS oncogene family |
| chr18_-_68433398 | 1.22 |
ENSMUST00000042852.7
|
Fam210a
|
family with sequence similarity 210, member A |
| chr2_+_15060051 | 1.22 |
ENSMUST00000069870.11
ENSMUST00000239125.2 ENSMUST00000193836.3 |
Arl5b
|
ADP-ribosylation factor-like 5B |
| chr4_+_41465134 | 1.22 |
ENSMUST00000030154.7
|
Nudt2
|
nudix (nucleoside diphosphate linked moiety X)-type motif 2 |
| chr5_+_54155814 | 1.22 |
ENSMUST00000117661.9
|
Stim2
|
stromal interaction molecule 2 |
| chr5_+_3646355 | 1.21 |
ENSMUST00000195894.2
|
Pex1
|
peroxisomal biogenesis factor 1 |
| chr10_+_62897353 | 1.20 |
ENSMUST00000178684.3
ENSMUST00000020266.15 |
Pbld1
|
phenazine biosynthesis-like protein domain containing 1 |
| chr19_+_43770619 | 1.19 |
ENSMUST00000026208.6
|
Abcc2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr4_-_91288221 | 1.19 |
ENSMUST00000102799.10
|
Elavl2
|
ELAV like RNA binding protein 1 |
| chr9_+_46151994 | 1.18 |
ENSMUST00000034585.7
|
Apoa4
|
apolipoprotein A-IV |
| chr6_+_95094721 | 1.18 |
ENSMUST00000032107.10
ENSMUST00000119582.3 |
Kbtbd8
|
kelch repeat and BTB (POZ) domain containing 8 |
| chr1_-_78488795 | 1.18 |
ENSMUST00000170511.3
|
BC035947
|
cDNA sequence BC035947 |
| chrM_+_7006 | 1.17 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chrX_+_100419965 | 1.13 |
ENSMUST00000119080.8
|
Gjb1
|
gap junction protein, beta 1 |
| chr3_+_144276154 | 1.13 |
ENSMUST00000082437.11
|
Selenof
|
selenoprotein F |
| chr11_-_76969230 | 1.13 |
ENSMUST00000102494.8
|
Nsrp1
|
nuclear speckle regulatory protein 1 |
| chr5_+_54155871 | 1.13 |
ENSMUST00000201469.4
|
Stim2
|
stromal interaction molecule 2 |
| chr11_-_3881995 | 1.12 |
ENSMUST00000020710.11
ENSMUST00000109989.10 ENSMUST00000109991.8 ENSMUST00000109993.9 |
Tcn2
|
transcobalamin 2 |
| chr6_-_128415640 | 1.12 |
ENSMUST00000032508.11
|
Fkbp4
|
FK506 binding protein 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Cux1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 17.1 | GO:1903699 | tarsal gland development(GO:1903699) |
| 5.3 | 26.7 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 2.6 | 7.9 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 2.6 | 10.4 | GO:0030091 | protein repair(GO:0030091) |
| 2.6 | 7.7 | GO:0090420 | naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 1.7 | 14.0 | GO:0071569 | protein ufmylation(GO:0071569) |
| 1.7 | 5.2 | GO:1903000 | regulation of lipid transport across blood brain barrier(GO:1903000) positive regulation of lipid transport across blood brain barrier(GO:1903002) |
| 1.5 | 18.1 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 1.5 | 4.5 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 1.3 | 5.2 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 1.2 | 1.2 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 1.1 | 3.3 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 1.1 | 3.3 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.9 | 4.6 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.9 | 5.4 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.8 | 2.5 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.8 | 3.4 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.8 | 3.3 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) |
| 0.8 | 3.9 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.7 | 4.2 | GO:0030300 | regulation of intestinal cholesterol absorption(GO:0030300) |
| 0.7 | 18.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.7 | 2.0 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.7 | 7.3 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.6 | 4.4 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.6 | 2.9 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.6 | 5.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.6 | 2.9 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.6 | 1.2 | GO:1902958 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) |
| 0.6 | 5.8 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.6 | 3.9 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.6 | 2.2 | GO:0035962 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 0.5 | 5.5 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.5 | 2.5 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.5 | 7.3 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.5 | 1.9 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 0.5 | 2.4 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.5 | 8.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.5 | 2.8 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.4 | 1.8 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.4 | 2.9 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.4 | 2.8 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response(GO:0030969) mRNA splicing, via endonucleolytic cleavage and ligation(GO:0070054) mRNA endonucleolytic cleavage involved in unfolded protein response(GO:0070055) |
| 0.4 | 2.0 | GO:0014028 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) notochord formation(GO:0014028) |
| 0.4 | 1.2 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.4 | 1.2 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.4 | 1.4 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.4 | 2.1 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.3 | 2.4 | GO:0098705 | copper ion import across plasma membrane(GO:0098705) copper ion import into cell(GO:1902861) |
| 0.3 | 2.0 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 | 1.3 | GO:0080163 | regulation of protein serine/threonine phosphatase activity(GO:0080163) |
| 0.3 | 1.0 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.3 | 1.0 | GO:0042853 | glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) L-alanine catabolic process(GO:0042853) oxalic acid secretion(GO:0046724) |
| 0.3 | 4.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.3 | 0.9 | GO:1902868 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.3 | 4.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.3 | 0.9 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.3 | 2.7 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.3 | 2.3 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.3 | 0.9 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.3 | 1.4 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.3 | 0.8 | GO:0016132 | brassinosteroid metabolic process(GO:0016131) brassinosteroid biosynthetic process(GO:0016132) |
| 0.3 | 0.8 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.3 | 0.8 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) |
| 0.3 | 0.8 | GO:1904155 | DN2 thymocyte differentiation(GO:1904155) DN3 thymocyte differentiation(GO:1904156) |
| 0.2 | 2.7 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.2 | 1.2 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.2 | 4.5 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.2 | 0.7 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.2 | 0.9 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.2 | 0.9 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.2 | 0.9 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.2 | 0.6 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.2 | 1.9 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.2 | 4.5 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.2 | 2.9 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.2 | 1.4 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.2 | 0.6 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.2 | 8.5 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.2 | 0.6 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.2 | 0.4 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.2 | 1.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.2 | 0.7 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.2 | 1.1 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.2 | 1.6 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.2 | 1.4 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 | 1.7 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.2 | 1.3 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.2 | 0.5 | GO:2000845 | negative regulation of glucagon secretion(GO:0070093) positive regulation of testosterone secretion(GO:2000845) |
| 0.2 | 0.5 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.2 | 1.1 | GO:0036476 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.2 | 8.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.2 | 0.6 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.2 | 1.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 0.7 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.1 | 3.0 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 4.8 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 3.9 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.6 | GO:0072061 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) inner medullary collecting duct development(GO:0072061) |
| 0.1 | 1.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 2.0 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.4 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.1 | 4.0 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 1.0 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 1.0 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.5 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 0.4 | GO:1902623 | negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 0.6 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 0.6 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.4 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.5 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 1.3 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.3 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.1 | 0.6 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.1 | 0.6 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.1 | 1.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.5 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.1 | 0.5 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 0.5 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 1.3 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 0.8 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 0.8 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 2.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 1.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 1.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 1.3 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.1 | 4.1 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.1 | 6.0 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.1 | 0.3 | GO:0048372 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) positive regulation of cardiac ventricle development(GO:1904414) |
| 0.1 | 1.5 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.7 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.1 | 0.4 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.1 | 1.8 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.2 | GO:0001806 | type IV hypersensitivity(GO:0001806) |
| 0.1 | 0.6 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 1.0 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.3 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.1 | 0.5 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.8 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 1.0 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.1 | 0.6 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 0.3 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 | 5.6 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.1 | 0.2 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.1 | 0.5 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 1.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.9 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.1 | 4.0 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 1.3 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 | 1.5 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.2 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.1 | 0.6 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.8 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.7 | GO:2001028 | positive regulation of endothelial cell chemotaxis(GO:2001028) |
| 0.1 | 1.7 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.1 | 1.6 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 1.4 | GO:0042761 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 2.6 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 0.3 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.1 | 6.2 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 0.5 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.1 | 1.1 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.1 | 0.7 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 1.7 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.1 | 1.4 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.1 | 0.3 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.3 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.2 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.2 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 0.3 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.7 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.4 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.8 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.1 | GO:0033693 | peripheral nervous system axon regeneration(GO:0014012) neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.6 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 8.8 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.8 | GO:0044331 | cell-cell adhesion mediated by cadherin(GO:0044331) |
| 0.0 | 0.3 | GO:0036394 | amylase secretion(GO:0036394) |
| 0.0 | 0.5 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.1 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.7 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.1 | GO:1904092 | regulation of autophagic cell death(GO:1904092) negative regulation of autophagic cell death(GO:1904093) |
| 0.0 | 0.3 | GO:0060460 | left lung development(GO:0060459) left lung morphogenesis(GO:0060460) |
| 0.0 | 2.3 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.5 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 0.3 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.8 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 3.7 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.9 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.0 | 0.4 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 1.4 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 8.7 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.3 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.2 | GO:0014717 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.6 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.2 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.0 | 0.4 | GO:0002827 | positive regulation of T-helper 1 type immune response(GO:0002827) |
| 0.0 | 0.8 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.5 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.2 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.4 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 1.1 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 2.0 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 0.5 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.3 | GO:0060253 | negative regulation of glial cell proliferation(GO:0060253) |
| 0.0 | 0.4 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.2 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.6 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.1 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.2 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.5 | GO:0023019 | signal transduction involved in regulation of gene expression(GO:0023019) |
| 0.0 | 0.9 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.4 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.9 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 1.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.6 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.6 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 1.7 | GO:0032835 | glomerulus development(GO:0032835) |
| 0.0 | 0.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.7 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 1.5 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.5 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.0 | 0.1 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 | 0.7 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.8 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.2 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.2 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.3 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.5 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 1.7 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.1 | GO:0022601 | menstrual cycle phase(GO:0022601) |
| 0.0 | 0.2 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.2 | GO:0045794 | negative regulation of cell volume(GO:0045794) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.3 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 0.7 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.4 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.9 | GO:0014032 | neural crest cell development(GO:0014032) |
| 0.0 | 1.5 | GO:0014013 | regulation of gliogenesis(GO:0014013) |
| 0.0 | 0.5 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.0 | 0.1 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.0 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.0 | 0.3 | GO:0051646 | mitochondrion localization(GO:0051646) |
| 0.0 | 1.0 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.2 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0044317 | rod spherule(GO:0044317) |
| 1.0 | 5.2 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.9 | 2.8 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.5 | 4.4 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.5 | 5.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.5 | 3.6 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.5 | 2.9 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.5 | 3.7 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.4 | 3.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.4 | 6.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.3 | 4.8 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.3 | 27.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 2.7 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.2 | 6.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 0.7 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.2 | 1.6 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.2 | 0.6 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.2 | 0.9 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.2 | 3.6 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 0.6 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.2 | 0.7 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.2 | 2.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.2 | 0.8 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 0.6 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 0.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 1.6 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 13.7 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 0.5 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 1.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.7 | GO:0098533 | ATPase dependent transmembrane transport complex(GO:0098533) |
| 0.1 | 1.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 18.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 0.8 | GO:0033647 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.1 | 0.7 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 9.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 5.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.8 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 5.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 4.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 1.0 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.2 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.1 | 17.6 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 2.0 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.9 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 0.9 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 0.7 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 1.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.8 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 1.1 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 1.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.3 | GO:0002142 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.1 | 0.7 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.7 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 0.3 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 0.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.9 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 1.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.8 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 2.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.7 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.0 | 0.1 | GO:0002095 | caveolar macromolecular signaling complex(GO:0002095) |
| 0.0 | 77.0 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.7 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 1.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.6 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 3.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.9 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 1.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 2.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 3.3 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 1.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.3 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 1.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 2.0 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 2.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.9 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.7 | GO:0032421 | stereocilium bundle(GO:0032421) |
| 0.0 | 0.1 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 1.0 | GO:0030139 | endocytic vesicle(GO:0030139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 18.1 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 2.2 | 13.4 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 2.2 | 13.0 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 1.8 | 5.3 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 1.7 | 10.4 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 1.7 | 5.2 | GO:0046911 | metal chelating activity(GO:0046911) |
| 1.4 | 4.1 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 1.3 | 5.3 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 1.3 | 6.4 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 1.2 | 27.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 1.0 | 6.0 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 1.0 | 4.8 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.9 | 4.6 | GO:0004031 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) molybdenum ion binding(GO:0030151) |
| 0.9 | 6.3 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.9 | 3.5 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.8 | 2.5 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.8 | 3.4 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.8 | 2.3 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.8 | 5.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.7 | 4.4 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.7 | 2.1 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.7 | 2.0 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 0.6 | 2.4 | GO:0019862 | IgA binding(GO:0019862) |
| 0.6 | 13.2 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.6 | 4.5 | GO:0015185 | gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.5 | 3.3 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.5 | 2.0 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.5 | 12.6 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.5 | 2.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.5 | 1.9 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.5 | 1.9 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.5 | 1.4 | GO:0080023 | 3R-hydroxyacyl-CoA dehydratase activity(GO:0080023) |
| 0.5 | 2.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.4 | 3.0 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.4 | 9.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.4 | 5.4 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.4 | 1.2 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.4 | 2.7 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.4 | 2.6 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.4 | 2.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.3 | 3.8 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 5.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.3 | 4.4 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.3 | 1.3 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.3 | 3.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.3 | 8.1 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.3 | 10.0 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 1.6 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.3 | 1.9 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.3 | 1.2 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.3 | 6.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.3 | 0.9 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.3 | 0.8 | GO:0047598 | sterol delta7 reductase activity(GO:0009918) 7-dehydrocholesterol reductase activity(GO:0047598) |
| 0.3 | 0.6 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.3 | 0.8 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.3 | 0.8 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.3 | 10.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.3 | 0.8 | GO:0004368 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.3 | 1.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.3 | 6.8 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.3 | 2.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 4.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.2 | 2.2 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.2 | 0.7 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.2 | 5.9 | GO:0001848 | complement binding(GO:0001848) |
| 0.2 | 1.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.2 | 0.2 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.2 | 2.4 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.2 | 3.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 1.6 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.2 | 0.8 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.2 | 5.7 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 2.9 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 0.6 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.2 | 3.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 0.4 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.2 | 0.9 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.2 | 2.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 0.7 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.2 | 0.7 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.2 | 0.9 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 1.0 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.2 | 0.5 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.2 | 2.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 3.0 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.4 | GO:0097604 | temperature-gated cation channel activity(GO:0097604) |
| 0.1 | 0.5 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 1.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 1.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 4.9 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.4 | GO:0004771 | sterol esterase activity(GO:0004771) retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 2.2 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 2.5 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 1.4 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 1.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 2.8 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.6 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.5 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.7 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.6 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 2.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 11.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.7 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 4.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.3 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.1 | 0.4 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 1.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 0.9 | GO:0043121 | neurotrophin binding(GO:0043121) nerve growth factor binding(GO:0048406) |
| 0.1 | 2.6 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 1.2 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 0.9 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.6 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.3 | GO:0038100 | nodal binding(GO:0038100) activin receptor binding(GO:0070697) |
| 0.1 | 0.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.5 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 1.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.6 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 1.2 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 1.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.7 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.3 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.6 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.5 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 4.4 | GO:0038024 | cargo receptor activity(GO:0038024) |
| 0.1 | 2.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 1.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 3.4 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.2 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 1.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.7 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 2.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.6 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 2.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.9 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.4 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 2.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.4 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.3 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 2.1 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.0 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 2.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 2.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 5.1 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 1.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.0 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 1.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.1 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 3.1 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.5 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.3 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.0 | 0.1 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.0 | 0.1 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 2.4 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 2.0 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 1.1 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 1.7 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.6 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 1.2 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.6 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.9 | GO:0016810 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds(GO:0016810) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 4.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 5.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 4.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 4.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 5.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 17.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 3.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 2.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 3.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 3.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.3 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 26.7 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.4 | 10.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 4.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.3 | 4.7 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.3 | 6.1 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.3 | 3.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 4.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 6.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 4.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 4.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 4.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 7.7 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.2 | 3.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.2 | 2.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 6.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 2.0 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 5.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 4.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 2.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 1.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 3.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 0.8 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 2.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 2.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 2.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 4.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 0.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.9 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 0.9 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 1.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 2.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 3.3 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 1.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 3.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 3.1 | REACTOME UNFOLDED PROTEIN RESPONSE | Genes involved in Unfolded Protein Response |
| 0.0 | 4.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.3 | REACTOME NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Neurotransmitter Release Cycle |
| 0.0 | 0.7 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 2.1 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 1.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 5.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.9 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 2.0 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |