Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
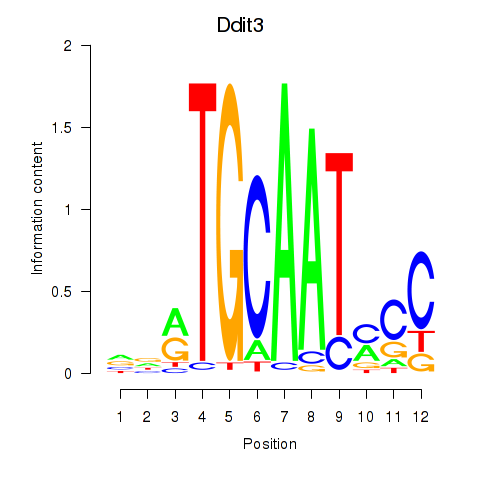
Results for Ddit3
Z-value: 0.53
Transcription factors associated with Ddit3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ddit3
|
ENSMUSG00000025408.16 | DNA-damage inducible transcript 3 |
|
Ddit3
|
ENSMUSG00000116429.2 | DNA-damage inducible transcript 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ddit3 | mm39_v1_chr10_+_127126643_127126698 | -0.38 | 2.1e-02 | Click! |
Activity profile of Ddit3 motif
Sorted Z-values of Ddit3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_105858432 | 2.29 |
ENSMUST00000161289.2
|
Ces4a
|
carboxylesterase 4A |
| chr2_+_119181703 | 2.23 |
ENSMUST00000028780.4
|
Chac1
|
ChaC, cation transport regulator 1 |
| chr14_-_67106037 | 1.46 |
ENSMUST00000022629.9
|
Dpysl2
|
dihydropyrimidinase-like 2 |
| chr7_-_46365108 | 1.29 |
ENSMUST00000006956.9
ENSMUST00000210913.2 |
Saa3
|
serum amyloid A 3 |
| chr5_+_33879018 | 0.76 |
ENSMUST00000201437.4
ENSMUST00000067150.14 ENSMUST00000169212.9 ENSMUST00000114411.9 ENSMUST00000164207.10 ENSMUST00000087820.8 |
Fgfr3
|
fibroblast growth factor receptor 3 |
| chr11_-_95966407 | 0.75 |
ENSMUST00000107686.8
ENSMUST00000107684.2 |
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
| chr11_+_70104929 | 0.72 |
ENSMUST00000094055.10
ENSMUST00000126296.8 ENSMUST00000136328.2 ENSMUST00000153993.3 |
Slc16a11
|
solute carrier family 16 (monocarboxylic acid transporters), member 11 |
| chr6_+_21215472 | 0.68 |
ENSMUST00000081542.6
|
Kcnd2
|
potassium voltage-gated channel, Shal-related family, member 2 |
| chr6_+_45036970 | 0.67 |
ENSMUST00000114641.8
|
Cntnap2
|
contactin associated protein-like 2 |
| chr6_+_90443293 | 0.66 |
ENSMUST00000203607.2
|
Klf15
|
Kruppel-like factor 15 |
| chr6_+_38511758 | 0.65 |
ENSMUST00000019833.5
|
Fmc1
|
formation of mitochondrial complex V assembly factor 1 |
| chr11_-_95966477 | 0.63 |
ENSMUST00000090541.12
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
| chr18_+_39126178 | 0.62 |
ENSMUST00000097593.9
|
Arhgap26
|
Rho GTPase activating protein 26 |
| chr8_-_71085097 | 0.60 |
ENSMUST00000110103.2
|
Gdf15
|
growth differentiation factor 15 |
| chr11_+_70104736 | 0.60 |
ENSMUST00000171032.8
|
Slc16a11
|
solute carrier family 16 (monocarboxylic acid transporters), member 11 |
| chr1_-_51955054 | 0.54 |
ENSMUST00000018561.14
ENSMUST00000114537.9 |
Myo1b
|
myosin IB |
| chr8_+_85807566 | 0.54 |
ENSMUST00000140621.2
|
Wdr83os
|
WD repeat domain 83 opposite strand |
| chr1_-_51955126 | 0.53 |
ENSMUST00000046390.14
|
Myo1b
|
myosin IB |
| chr3_-_91990439 | 0.51 |
ENSMUST00000058150.8
|
Lor
|
loricrin |
| chr8_+_85807369 | 0.50 |
ENSMUST00000079764.14
|
Wdr83os
|
WD repeat domain 83 opposite strand |
| chr11_+_95152355 | 0.50 |
ENSMUST00000021242.5
|
Tac4
|
tachykinin 4 |
| chr7_+_43362823 | 0.49 |
ENSMUST00000206554.2
|
Klk13
|
kallikrein related-peptidase 13 |
| chr9_+_20556088 | 0.49 |
ENSMUST00000162303.8
ENSMUST00000161486.8 |
Ubl5
|
ubiquitin-like 5 |
| chr1_-_5089564 | 0.48 |
ENSMUST00000002533.15
|
Rgs20
|
regulator of G-protein signaling 20 |
| chr18_+_37440497 | 0.47 |
ENSMUST00000056712.4
|
Pcdhb4
|
protocadherin beta 4 |
| chr7_+_126376099 | 0.44 |
ENSMUST00000038614.12
ENSMUST00000170882.8 ENSMUST00000106359.2 ENSMUST00000106357.8 ENSMUST00000145762.8 |
Ypel3
|
yippee like 3 |
| chr8_+_46080840 | 0.43 |
ENSMUST00000135336.9
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr7_+_126376353 | 0.43 |
ENSMUST00000106356.2
|
Ypel3
|
yippee like 3 |
| chr9_+_20556147 | 0.43 |
ENSMUST00000161882.8
|
Ubl5
|
ubiquitin-like 5 |
| chr18_-_35760260 | 0.42 |
ENSMUST00000025212.8
|
Slc23a1
|
solute carrier family 23 (nucleobase transporters), member 1 |
| chr5_-_44139121 | 0.42 |
ENSMUST00000199894.2
|
Fgfbp1
|
fibroblast growth factor binding protein 1 |
| chr5_+_137551790 | 0.42 |
ENSMUST00000136565.8
ENSMUST00000149292.8 ENSMUST00000125489.2 |
Actl6b
|
actin-like 6B |
| chr11_-_103109247 | 0.41 |
ENSMUST00000103076.2
|
Spata32
|
spermatogenesis associated 32 |
| chr3_+_29568055 | 0.38 |
ENSMUST00000140288.2
|
Egfem1
|
EGF-like and EMI domain containing 1 |
| chr2_-_90735171 | 0.38 |
ENSMUST00000005647.4
|
Ndufs3
|
NADH:ubiquinone oxidoreductase core subunit S3 |
| chr11_+_60619224 | 0.38 |
ENSMUST00000018743.5
|
Mief2
|
mitochondrial elongation factor 2 |
| chr7_+_126376319 | 0.38 |
ENSMUST00000132643.2
|
Ypel3
|
yippee like 3 |
| chr5_-_44139099 | 0.37 |
ENSMUST00000061299.9
|
Fgfbp1
|
fibroblast growth factor binding protein 1 |
| chr11_+_110289941 | 0.37 |
ENSMUST00000020949.12
ENSMUST00000100260.2 |
Map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr9_+_65278979 | 0.37 |
ENSMUST00000239433.2
|
Ubap1l
|
ubiquitin-associated protein 1-like |
| chr7_+_30475819 | 0.37 |
ENSMUST00000041703.10
|
Dmkn
|
dermokine |
| chr8_+_12965876 | 0.37 |
ENSMUST00000110876.9
ENSMUST00000110879.9 |
Mcf2l
|
mcf.2 transforming sequence-like |
| chr14_+_66208253 | 0.37 |
ENSMUST00000138191.8
|
Clu
|
clusterin |
| chr19_+_47853981 | 0.37 |
ENSMUST00000120645.8
|
Gsto2
|
glutathione S-transferase omega 2 |
| chr17_-_25459086 | 0.36 |
ENSMUST00000038973.7
ENSMUST00000115154.11 |
Gnptg
|
N-acetylglucosamine-1-phosphotransferase, gamma subunit |
| chr5_+_137551774 | 0.36 |
ENSMUST00000136088.8
ENSMUST00000139395.8 |
Actl6b
|
actin-like 6B |
| chr11_+_110888313 | 0.35 |
ENSMUST00000106635.2
|
Kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr14_+_53913598 | 0.35 |
ENSMUST00000103662.6
|
Trav9-4
|
T cell receptor alpha variable 9-4 |
| chr15_+_102922247 | 0.35 |
ENSMUST00000001709.3
|
Hoxc5
|
homeobox C5 |
| chr8_-_100143029 | 0.34 |
ENSMUST00000155527.8
ENSMUST00000142129.8 ENSMUST00000093249.11 ENSMUST00000142475.3 ENSMUST00000128860.8 |
Cdh8
|
cadherin 8 |
| chr18_+_39126325 | 0.33 |
ENSMUST00000137497.9
|
Arhgap26
|
Rho GTPase activating protein 26 |
| chr17_-_15163362 | 0.33 |
ENSMUST00000238668.2
ENSMUST00000228330.2 |
Wdr27
|
WD repeat domain 27 |
| chr19_+_47854012 | 0.32 |
ENSMUST00000237514.2
|
Gsto2
|
glutathione S-transferase omega 2 |
| chr7_-_103845192 | 0.32 |
ENSMUST00000106859.3
|
Olfr649
|
olfactory receptor 649 |
| chr5_+_32616566 | 0.32 |
ENSMUST00000202078.2
|
Ppp1cb
|
protein phosphatase 1 catalytic subunit beta |
| chr8_-_111808488 | 0.32 |
ENSMUST00000188466.7
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr1_+_36753607 | 0.32 |
ENSMUST00000208994.2
|
4933424G06Rik
|
RIKEN cDNA 4933424G06 gene |
| chr7_+_144450260 | 0.32 |
ENSMUST00000033389.7
ENSMUST00000207229.2 |
Fgf15
|
fibroblast growth factor 15 |
| chr11_+_54194624 | 0.31 |
ENSMUST00000093106.12
|
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr10_-_95158827 | 0.31 |
ENSMUST00000220279.2
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr14_+_66208498 | 0.30 |
ENSMUST00000128539.8
|
Clu
|
clusterin |
| chr7_+_99659121 | 0.30 |
ENSMUST00000107084.8
|
Chrdl2
|
chordin-like 2 |
| chr7_+_30681287 | 0.30 |
ENSMUST00000128384.3
|
Fam187b
|
family with sequence similarity 187, member B |
| chrX_+_165021919 | 0.30 |
ENSMUST00000060210.14
ENSMUST00000112233.8 |
Gpm6b
|
glycoprotein m6b |
| chr2_-_121637469 | 0.30 |
ENSMUST00000110592.2
|
Frmd5
|
FERM domain containing 5 |
| chr5_-_129916283 | 0.30 |
ENSMUST00000094280.4
|
Chchd2
|
coiled-coil-helix-coiled-coil-helix domain containing 2 |
| chr19_+_47853954 | 0.29 |
ENSMUST00000235896.2
|
Gsto2
|
glutathione S-transferase omega 2 |
| chr9_+_18848418 | 0.29 |
ENSMUST00000218385.2
|
Olfr832
|
olfactory receptor 832 |
| chr15_-_12549963 | 0.29 |
ENSMUST00000189324.2
|
Pdzd2
|
PDZ domain containing 2 |
| chr9_-_45817666 | 0.29 |
ENSMUST00000161187.8
|
Rnf214
|
ring finger protein 214 |
| chr5_-_53864595 | 0.29 |
ENSMUST00000200691.4
|
Cckar
|
cholecystokinin A receptor |
| chr12_-_87149124 | 0.28 |
ENSMUST00000110176.2
|
Ngb
|
neuroglobin |
| chr7_+_120682472 | 0.28 |
ENSMUST00000171880.3
ENSMUST00000047025.15 ENSMUST00000170106.2 ENSMUST00000168311.8 |
Otoa
|
otoancorin |
| chr19_+_4889394 | 0.28 |
ENSMUST00000088653.3
|
Ccdc87
|
coiled-coil domain containing 87 |
| chrX_-_105528503 | 0.28 |
ENSMUST00000138724.8
ENSMUST00000149331.2 |
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chr2_+_140012560 | 0.28 |
ENSMUST00000044825.5
|
Ndufaf5
|
NADH:ubiquinone oxidoreductase complex assembly factor 5 |
| chr1_-_37469037 | 0.28 |
ENSMUST00000027286.7
|
Coa5
|
cytochrome C oxidase assembly factor 5 |
| chr2_+_109522781 | 0.27 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr19_+_8975249 | 0.27 |
ENSMUST00000236390.2
|
Ahnak
|
AHNAK nucleoprotein (desmoyokin) |
| chr9_+_57143839 | 0.27 |
ENSMUST00000093837.5
|
Trcg1
|
taste receptor cell gene 1 |
| chr8_+_127025265 | 0.27 |
ENSMUST00000108759.3
|
Slc35f3
|
solute carrier family 35, member F3 |
| chr2_+_147203845 | 0.27 |
ENSMUST00000126068.9
|
Pax1
|
paired box 1 |
| chr2_-_153712996 | 0.26 |
ENSMUST00000028982.5
|
Sun5
|
Sad1 and UNC84 domain containing 5 |
| chr8_-_85807308 | 0.26 |
ENSMUST00000093357.12
|
Wdr83
|
WD repeat domain containing 83 |
| chr10_-_95159410 | 0.25 |
ENSMUST00000217809.2
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr7_+_102289455 | 0.25 |
ENSMUST00000098221.2
|
Olfr554
|
olfactory receptor 554 |
| chr2_+_22959223 | 0.25 |
ENSMUST00000114523.10
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr3_+_108646974 | 0.25 |
ENSMUST00000133931.9
|
Aknad1
|
AKNA domain containing 1 |
| chr4_-_56990306 | 0.25 |
ENSMUST00000053681.6
|
Frrs1l
|
ferric-chelate reductase 1 like |
| chr4_+_154921916 | 0.25 |
ENSMUST00000207854.2
|
Ttc34
|
tetratricopeptide repeat domain 34 |
| chr3_-_151541603 | 0.25 |
ENSMUST00000106126.2
|
Ptgfr
|
prostaglandin F receptor |
| chr19_+_46750016 | 0.25 |
ENSMUST00000099373.12
ENSMUST00000077666.6 |
Cnnm2
|
cyclin M2 |
| chr2_+_90229027 | 0.24 |
ENSMUST00000216111.3
|
Olfr1274
|
olfactory receptor 1274 |
| chr6_+_18170686 | 0.24 |
ENSMUST00000045706.12
|
Cftr
|
cystic fibrosis transmembrane conductance regulator |
| chr11_-_87716849 | 0.24 |
ENSMUST00000103177.10
|
Lpo
|
lactoperoxidase |
| chr7_+_120276801 | 0.23 |
ENSMUST00000208454.2
ENSMUST00000060175.8 |
Mosmo
|
modulator of smoothened |
| chr17_+_26633794 | 0.23 |
ENSMUST00000182897.8
ENSMUST00000183077.8 ENSMUST00000053020.8 |
Neurl1b
|
neuralized E3 ubiquitin protein ligase 1B |
| chr7_-_42376441 | 0.23 |
ENSMUST00000166837.2
|
Gm17067
|
predicted gene 17067 |
| chr2_-_172296662 | 0.23 |
ENSMUST00000161334.2
|
Gcnt7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr15_-_43733389 | 0.23 |
ENSMUST00000067469.6
|
Tmem74
|
transmembrane protein 74 |
| chr4_-_117622747 | 0.23 |
ENSMUST00000062747.6
|
Klf17
|
Kruppel-like factor 17 |
| chrX_-_166638057 | 0.22 |
ENSMUST00000238211.2
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr5_-_92511136 | 0.22 |
ENSMUST00000077820.6
|
Cxcl11
|
chemokine (C-X-C motif) ligand 11 |
| chr17_-_5468938 | 0.22 |
ENSMUST00000189788.2
|
Ldhal6b
|
lactate dehydrogenase A-like 6B |
| chr2_+_36263531 | 0.22 |
ENSMUST00000072114.4
ENSMUST00000217511.2 |
Olfr338
|
olfactory receptor 338 |
| chr6_-_125143425 | 0.22 |
ENSMUST00000117757.9
ENSMUST00000073605.15 |
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr3_-_153611775 | 0.22 |
ENSMUST00000005630.11
|
Msh4
|
mutS homolog 4 |
| chr7_-_23206631 | 0.22 |
ENSMUST00000227713.2
|
Vmn1r167
|
vomeronasal 1 receptor 167 |
| chr3_-_58433313 | 0.22 |
ENSMUST00000029385.9
|
Serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr5_-_108922819 | 0.22 |
ENSMUST00000200159.2
ENSMUST00000212212.2 |
Rnf212
|
ring finger protein 212 |
| chr4_+_112470794 | 0.22 |
ENSMUST00000058791.14
ENSMUST00000186969.2 |
Skint2
|
selection and upkeep of intraepithelial T cells 2 |
| chr2_-_91025208 | 0.22 |
ENSMUST00000111355.8
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr4_-_101701220 | 0.22 |
ENSMUST00000106919.8
|
Pramel17
|
PRAME like 17 |
| chr1_-_126420420 | 0.22 |
ENSMUST00000162877.8
|
Nckap5
|
NCK-associated protein 5 |
| chr8_-_68270870 | 0.21 |
ENSMUST00000059374.5
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chrX_+_102465616 | 0.21 |
ENSMUST00000182089.2
|
Gm26992
|
predicted gene, 26992 |
| chr13_+_34918820 | 0.21 |
ENSMUST00000039605.8
|
Fam50b
|
family with sequence similarity 50, member B |
| chr14_+_66208613 | 0.21 |
ENSMUST00000144619.2
|
Clu
|
clusterin |
| chr12_+_78738425 | 0.21 |
ENSMUST00000218697.2
ENSMUST00000219551.2 ENSMUST00000220396.2 |
Fam71d
|
family with sequence similarity 71, member D |
| chr6_-_125142539 | 0.20 |
ENSMUST00000183272.2
ENSMUST00000182052.8 ENSMUST00000182277.2 |
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr6_+_17463748 | 0.20 |
ENSMUST00000115443.8
|
Met
|
met proto-oncogene |
| chrX_-_59193003 | 0.20 |
ENSMUST00000033478.5
ENSMUST00000101531.10 |
Mcf2
|
mcf.2 transforming sequence |
| chr14_+_26789345 | 0.20 |
ENSMUST00000226105.2
|
Il17rd
|
interleukin 17 receptor D |
| chr9_-_121324705 | 0.20 |
ENSMUST00000216138.2
|
Cck
|
cholecystokinin |
| chr16_-_45544960 | 0.20 |
ENSMUST00000096057.5
|
Tagln3
|
transgelin 3 |
| chr12_+_78738288 | 0.19 |
ENSMUST00000077968.5
|
Fam71d
|
family with sequence similarity 71, member D |
| chr11_-_6015736 | 0.19 |
ENSMUST00000002817.12
ENSMUST00000109813.9 ENSMUST00000090443.10 |
Camk2b
|
calcium/calmodulin-dependent protein kinase II, beta |
| chr18_+_37063237 | 0.19 |
ENSMUST00000193839.6
ENSMUST00000070797.7 |
Pcdha1
|
protocadherin alpha 1 |
| chr5_-_53864874 | 0.19 |
ENSMUST00000031093.5
|
Cckar
|
cholecystokinin A receptor |
| chr14_+_53828314 | 0.19 |
ENSMUST00000103654.3
|
Trav9-2
|
T cell receptor alpha variable 9-2 |
| chr1_+_5658716 | 0.18 |
ENSMUST00000160777.8
ENSMUST00000239100.2 ENSMUST00000027038.11 |
Oprk1
|
opioid receptor, kappa 1 |
| chr17_+_69382668 | 0.18 |
ENSMUST00000223610.2
|
Epb41l3
|
erythrocyte membrane protein band 4.1 like 3 |
| chr19_+_9824919 | 0.18 |
ENSMUST00000179814.3
|
Scgb2a2
|
secretoglobin, family 2A, member 2 |
| chr4_+_129841340 | 0.18 |
ENSMUST00000128007.8
|
Spocd1
|
SPOC domain containing 1 |
| chr7_-_125681577 | 0.18 |
ENSMUST00000073935.7
|
Gsg1l
|
GSG1-like |
| chr2_-_91025441 | 0.18 |
ENSMUST00000002177.9
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chrX_-_74207359 | 0.18 |
ENSMUST00000101433.9
|
Smim9
|
small integral membrane protein 9 |
| chr17_-_15784582 | 0.18 |
ENSMUST00000147532.8
|
Prdm9
|
PR domain containing 9 |
| chr2_-_90054837 | 0.18 |
ENSMUST00000213994.3
|
Olfr1506
|
olfactory receptor 1506 |
| chr14_-_51287620 | 0.17 |
ENSMUST00000076106.4
|
Rnase11
|
ribonuclease, RNase A family, 11 (non-active) |
| chr9_-_58009508 | 0.17 |
ENSMUST00000119665.2
|
Ccdc33
|
coiled-coil domain containing 33 |
| chr18_+_37554471 | 0.17 |
ENSMUST00000053073.6
|
Pcdhb11
|
protocadherin beta 11 |
| chr3_-_152045986 | 0.17 |
ENSMUST00000199397.2
ENSMUST00000199334.5 ENSMUST00000068243.11 ENSMUST00000073089.13 |
Miga1
|
mitoguardin 1 |
| chr6_+_42377172 | 0.17 |
ENSMUST00000057398.4
|
Tas2r143
|
taste receptor, type 2, member 143 |
| chr9_-_121324744 | 0.17 |
ENSMUST00000035120.6
|
Cck
|
cholecystokinin |
| chr15_-_98674910 | 0.16 |
ENSMUST00000228594.2
ENSMUST00000226846.2 |
Wnt10b
|
wingless-type MMTV integration site family, member 10B |
| chr7_+_84178162 | 0.16 |
ENSMUST00000180387.3
|
Gm2115
|
predicted gene 2115 |
| chr6_+_116528102 | 0.16 |
ENSMUST00000122096.3
|
Eif4a3l2
|
eukaryotic translation initiation factor 4A3 like 2 |
| chr14_+_54183465 | 0.16 |
ENSMUST00000197130.5
ENSMUST00000103677.3 |
Trdv2-1
|
T cell receptor delta variable 2-1 |
| chr1_-_161807205 | 0.16 |
ENSMUST00000162676.2
|
4930558K02Rik
|
RIKEN cDNA 4930558K02 gene |
| chr6_+_120070307 | 0.16 |
ENSMUST00000112711.9
|
Ninj2
|
ninjurin 2 |
| chr9_+_104446682 | 0.16 |
ENSMUST00000057742.15
|
Cpne4
|
copine IV |
| chr3_+_82287380 | 0.16 |
ENSMUST00000195793.2
|
Map9
|
microtubule-associated protein 9 |
| chr5_-_131567526 | 0.16 |
ENSMUST00000161374.8
|
Auts2
|
autism susceptibility candidate 2 |
| chr4_+_101407608 | 0.15 |
ENSMUST00000094953.11
ENSMUST00000106933.2 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr9_+_107957621 | 0.15 |
ENSMUST00000035211.14
|
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr2_+_121787131 | 0.15 |
ENSMUST00000110574.8
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr7_+_134870237 | 0.15 |
ENSMUST00000210697.2
ENSMUST00000097983.5 |
Nps
|
neuropeptide S |
| chr9_-_108305683 | 0.15 |
ENSMUST00000076592.4
|
Iho1
|
interactor of HORMAD1 1 |
| chr5_-_113229445 | 0.15 |
ENSMUST00000131708.2
ENSMUST00000117143.8 ENSMUST00000119627.8 |
Crybb3
|
crystallin, beta B3 |
| chr7_-_104677667 | 0.15 |
ENSMUST00000215899.2
ENSMUST00000214318.3 |
Olfr675
|
olfactory receptor 675 |
| chr19_-_5156995 | 0.15 |
ENSMUST00000237438.2
ENSMUST00000236149.2 ENSMUST00000025804.7 |
Rab1b
|
RAB1B, member RAS oncogene family |
| chr9_+_38312994 | 0.15 |
ENSMUST00000214648.3
ENSMUST00000056364.3 |
Olfr147
|
olfactory receptor 147 |
| chr7_-_79443536 | 0.15 |
ENSMUST00000032760.6
|
Mesp1
|
mesoderm posterior 1 |
| chr19_-_12209960 | 0.15 |
ENSMUST00000207710.3
|
Olfr1432
|
olfactory receptor 1432 |
| chr6_+_88701578 | 0.14 |
ENSMUST00000150180.4
ENSMUST00000163271.8 |
Mgll
|
monoglyceride lipase |
| chr2_+_90152544 | 0.14 |
ENSMUST00000214797.3
|
Olfr1565
|
olfactory receptor 1565 |
| chr2_-_91025380 | 0.14 |
ENSMUST00000111356.8
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr5_+_32616187 | 0.14 |
ENSMUST00000015100.15
|
Ppp1cb
|
protein phosphatase 1 catalytic subunit beta |
| chr2_+_152446244 | 0.14 |
ENSMUST00000058086.6
|
Defb36
|
defensin beta 36 |
| chrX_+_51880056 | 0.14 |
ENSMUST00000101588.2
|
Ccdc160
|
coiled-coil domain containing 160 |
| chr5_-_129856237 | 0.14 |
ENSMUST00000118268.9
|
Psph
|
phosphoserine phosphatase |
| chr2_+_131048998 | 0.14 |
ENSMUST00000153097.3
|
Ap5s1
|
adaptor-related protein 5 complex, sigma 1 subunit |
| chr18_-_60724855 | 0.14 |
ENSMUST00000056533.9
|
Myoz3
|
myozenin 3 |
| chr14_-_70890521 | 0.13 |
ENSMUST00000062629.5
|
Npm2
|
nucleophosmin/nucleoplasmin 2 |
| chr2_-_88837317 | 0.13 |
ENSMUST00000216271.3
ENSMUST00000214809.3 |
Olfr1215
|
olfactory receptor 1215 |
| chr1_+_187995096 | 0.13 |
ENSMUST00000060479.14
|
Ush2a
|
usherin |
| chr17_+_69382694 | 0.13 |
ENSMUST00000225062.2
|
Epb41l3
|
erythrocyte membrane protein band 4.1 like 3 |
| chr14_+_32972324 | 0.13 |
ENSMUST00000131086.3
|
Arhgap22
|
Rho GTPase activating protein 22 |
| chrY_-_35085749 | 0.13 |
ENSMUST00000180170.2
|
Gm20855
|
predicted gene, 20855 |
| chr9_+_107957640 | 0.13 |
ENSMUST00000162886.2
|
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr7_+_23811739 | 0.13 |
ENSMUST00000120006.8
ENSMUST00000005413.4 |
Zfp112
|
zinc finger protein 112 |
| chr6_+_88701810 | 0.13 |
ENSMUST00000089449.5
|
Mgll
|
monoglyceride lipase |
| chr9_-_40015750 | 0.13 |
ENSMUST00000213858.2
|
Olfr984
|
olfactory receptor 984 |
| chr6_+_71299758 | 0.12 |
ENSMUST00000065248.9
|
Cd8b1
|
CD8 antigen, beta chain 1 |
| chr6_+_41510925 | 0.12 |
ENSMUST00000103285.2
|
Trbj1-2
|
T cell receptor beta joining 1-2 |
| chr12_+_53294940 | 0.12 |
ENSMUST00000223358.3
|
Npas3
|
neuronal PAS domain protein 3 |
| chr3_-_58729732 | 0.12 |
ENSMUST00000191233.4
|
Mindy4b-ps
|
MINDY lysine 48 deubiquitinase 4B, pseudogene |
| chr18_+_37466877 | 0.12 |
ENSMUST00000194655.2
ENSMUST00000061717.4 |
Pcdhb6
|
protocadherin beta 6 |
| chr14_-_47805861 | 0.12 |
ENSMUST00000228784.2
ENSMUST00000042988.7 |
Atg14
|
autophagy related 14 |
| chr1_-_39616445 | 0.12 |
ENSMUST00000062525.11
|
Rnf149
|
ring finger protein 149 |
| chr8_-_68270936 | 0.12 |
ENSMUST00000120071.8
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr7_+_6392535 | 0.12 |
ENSMUST00000207465.2
ENSMUST00000208338.2 |
Zfp28
|
zinc finger protein 28 |
| chr6_-_57821483 | 0.12 |
ENSMUST00000226191.2
|
Vmn1r21
|
vomeronasal 1 receptor 21 |
| chr1_+_34160331 | 0.11 |
ENSMUST00000183006.5
|
Dst
|
dystonin |
| chr18_+_42408418 | 0.11 |
ENSMUST00000091920.6
ENSMUST00000046972.14 ENSMUST00000236240.2 |
Rbm27
|
RNA binding motif protein 27 |
| chr4_+_151081538 | 0.11 |
ENSMUST00000030803.2
|
Uts2
|
urotensin 2 |
| chr2_-_91025492 | 0.11 |
ENSMUST00000111354.2
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr16_+_4964849 | 0.11 |
ENSMUST00000165810.2
ENSMUST00000230616.2 |
Sec14l5
|
SEC14-like lipid binding 5 |
| chrY_+_79332266 | 0.11 |
ENSMUST00000178063.2
|
Gm20916
|
predicted gene, 20916 |
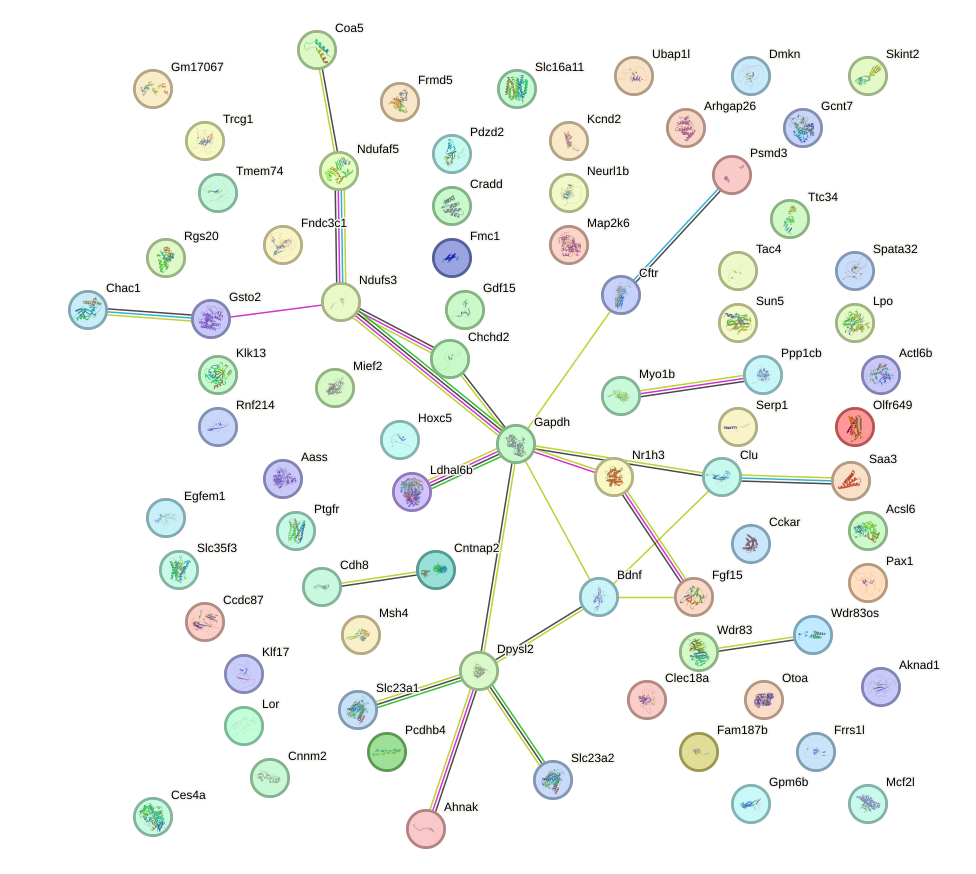
Network of associatons between targets according to the STRING database.
First level regulatory network of Ddit3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0021762 | substantia nigra development(GO:0021762) alveolar secondary septum development(GO:0061144) |
| 0.2 | 0.7 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.2 | 2.2 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 0.9 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 0.4 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.4 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 | 1.2 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 0.4 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.1 | 0.7 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 1.0 | GO:0090487 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 0.1 | 0.4 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.3 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.1 | 0.3 | GO:0061193 | taste bud development(GO:0061193) |
| 0.1 | 0.5 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.1 | 0.2 | GO:1902161 | transepithelial water transport(GO:0035377) positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) positive regulation of establishment of Sertoli cell barrier(GO:1904446) negative regulation of type B pancreatic cell development(GO:2000077) |
| 0.1 | 0.8 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 0.4 | GO:0035822 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 0.1 | 0.5 | GO:0090274 | reduction of food intake in response to dietary excess(GO:0002023) positive regulation of somatostatin secretion(GO:0090274) |
| 0.1 | 0.1 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.1 | 0.2 | GO:0060466 | activation of meiosis involved in egg activation(GO:0060466) |
| 0.1 | 0.3 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 1.9 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.1 | 0.3 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.1 | 0.2 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.1 | 0.3 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.1 | 0.3 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.1 | 1.3 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.1 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.0 | 0.3 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.3 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.3 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.0 | 1.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.7 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.3 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) |
| 0.0 | 0.1 | GO:0033967 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.5 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.3 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.0 | 0.2 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.0 | 0.3 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.0 | 0.2 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.6 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.4 | GO:0032308 | positive regulation of prostaglandin secretion(GO:0032308) |
| 0.0 | 0.1 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.0 | 0.1 | GO:0002849 | positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.0 | 0.2 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.2 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.1 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.0 | 0.4 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.0 | 0.4 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.7 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.6 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.6 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.3 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.2 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.1 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.5 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.3 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 0.1 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.0 | GO:0035874 | amiloride transport(GO:0015898) cellular response to copper ion starvation(GO:0035874) response to azide(GO:0097184) cellular response to azide(GO:0097185) |
| 0.0 | 0.0 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.4 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.2 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 0.9 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.1 | 0.5 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.1 | GO:0097632 | extrinsic component of pre-autophagosomal structure membrane(GO:0097632) |
| 0.0 | 0.2 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.0 | 0.2 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 1.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.0 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.8 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.5 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.7 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.3 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 1.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.2 | 1.3 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.4 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.1 | 1.0 | GO:0050610 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.1 | 0.7 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.7 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.1 | 0.4 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.1 | 0.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.3 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.4 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 0.3 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.1 | 0.2 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 1.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.9 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 1.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.0 | 0.1 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.9 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.2 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.6 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 1.3 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.0 | GO:0052597 | diamine oxidase activity(GO:0052597) histamine oxidase activity(GO:0052598) methylputrescine oxidase activity(GO:0052599) propane-1,3-diamine oxidase activity(GO:0052600) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.9 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 2.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.8 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 1.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |