Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
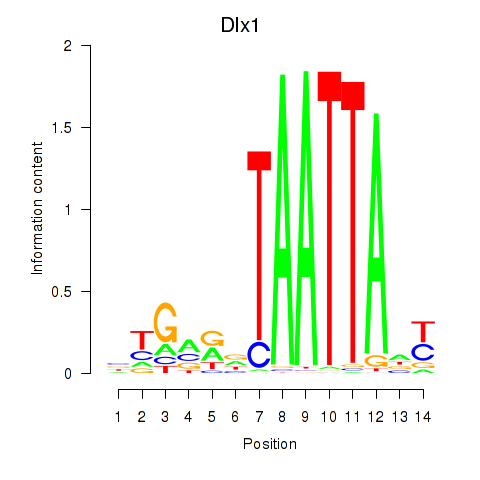
Results for Dlx1
Z-value: 1.20
Transcription factors associated with Dlx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Dlx1
|
ENSMUSG00000041911.4 | distal-less homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dlx1 | mm39_v1_chr2_+_71359000_71359027 | 0.31 | 6.7e-02 | Click! |
Activity profile of Dlx1 motif
Sorted Z-values of Dlx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_41121979 | 19.09 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr11_+_87684299 | 12.07 |
ENSMUST00000020779.11
|
Mpo
|
myeloperoxidase |
| chr3_-_106126794 | 9.95 |
ENSMUST00000082219.6
|
Chil4
|
chitinase-like 4 |
| chr14_+_80237691 | 9.49 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr1_-_132318039 | 9.07 |
ENSMUST00000132435.2
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chrX_+_149330371 | 8.86 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr10_+_115653152 | 8.21 |
ENSMUST00000080630.11
ENSMUST00000179196.3 ENSMUST00000035563.15 |
Tspan8
|
tetraspanin 8 |
| chr2_+_84818538 | 6.88 |
ENSMUST00000028466.12
|
Prg3
|
proteoglycan 3 |
| chr11_-_11920540 | 6.64 |
ENSMUST00000109653.8
|
Grb10
|
growth factor receptor bound protein 10 |
| chr2_+_174292471 | 6.44 |
ENSMUST00000016399.6
|
Tubb1
|
tubulin, beta 1 class VI |
| chr7_-_133304244 | 6.17 |
ENSMUST00000209636.2
ENSMUST00000153698.3 |
Uros
|
uroporphyrinogen III synthase |
| chr12_-_75678092 | 6.13 |
ENSMUST00000238938.2
|
Rplp2-ps1
|
ribosomal protein, large P2, pseudogene 1 |
| chr11_+_58808716 | 5.91 |
ENSMUST00000069941.13
|
Btnl10
|
butyrophilin-like 10 |
| chr8_+_94899292 | 5.46 |
ENSMUST00000034214.8
ENSMUST00000212806.2 |
Mt2
|
metallothionein 2 |
| chr11_+_58808830 | 5.21 |
ENSMUST00000020792.12
ENSMUST00000108818.4 |
Btnl10
|
butyrophilin-like 10 |
| chr19_-_40576782 | 5.17 |
ENSMUST00000176939.8
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr7_-_140597465 | 4.90 |
ENSMUST00000211330.2
|
Ifitm6
|
interferon induced transmembrane protein 6 |
| chr9_-_21874802 | 4.89 |
ENSMUST00000006397.7
|
Epor
|
erythropoietin receptor |
| chr10_+_20223516 | 4.79 |
ENSMUST00000169712.3
ENSMUST00000217608.2 |
Mtfr2
|
mitochondrial fission regulator 2 |
| chr11_-_106205320 | 4.08 |
ENSMUST00000167143.2
|
Cd79b
|
CD79B antigen |
| chr19_-_40576817 | 4.07 |
ENSMUST00000175932.2
ENSMUST00000176955.8 ENSMUST00000149476.3 |
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr10_-_62363217 | 4.03 |
ENSMUST00000160987.8
|
Srgn
|
serglycin |
| chr4_-_156340713 | 4.02 |
ENSMUST00000219393.2
|
Samd11
|
sterile alpha motif domain containing 11 |
| chr1_+_40478787 | 3.86 |
ENSMUST00000097772.10
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr7_+_27151838 | 3.56 |
ENSMUST00000108357.8
|
Blvrb
|
biliverdin reductase B (flavin reductase (NADPH)) |
| chr11_-_117671436 | 3.51 |
ENSMUST00000026659.10
ENSMUST00000127227.2 |
Tmc6
|
transmembrane channel-like gene family 6 |
| chr11_+_120499295 | 3.46 |
ENSMUST00000106194.8
ENSMUST00000106195.3 ENSMUST00000061309.5 |
Npb
|
neuropeptide B |
| chr2_-_164198427 | 3.45 |
ENSMUST00000109367.10
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr9_+_120400510 | 3.43 |
ENSMUST00000165532.3
|
Rpl14
|
ribosomal protein L14 |
| chr8_-_4829519 | 3.29 |
ENSMUST00000022945.9
|
Shcbp1
|
Shc SH2-domain binding protein 1 |
| chrX_+_139857640 | 3.23 |
ENSMUST00000112971.2
|
Atg4a
|
autophagy related 4A, cysteine peptidase |
| chr8_+_117648474 | 3.18 |
ENSMUST00000034205.5
ENSMUST00000212775.2 |
Cenpn
|
centromere protein N |
| chr14_+_26722319 | 3.13 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr2_+_84669739 | 3.09 |
ENSMUST00000146816.8
ENSMUST00000028469.14 |
Slc43a1
|
solute carrier family 43, member 1 |
| chr6_+_72074545 | 3.09 |
ENSMUST00000069994.11
ENSMUST00000114112.4 |
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr4_-_131802561 | 3.09 |
ENSMUST00000105970.8
ENSMUST00000105975.8 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr9_-_103357564 | 3.07 |
ENSMUST00000124310.5
|
Bfsp2
|
beaded filament structural protein 2, phakinin |
| chr13_+_52750883 | 3.04 |
ENSMUST00000055087.7
|
Syk
|
spleen tyrosine kinase |
| chr16_-_75706161 | 2.98 |
ENSMUST00000114239.9
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr3_-_14873406 | 2.95 |
ENSMUST00000181860.8
ENSMUST00000144327.3 |
Car1
|
carbonic anhydrase 1 |
| chr2_+_85809620 | 2.93 |
ENSMUST00000056849.3
|
Olfr1030
|
olfactory receptor 1030 |
| chr3_-_130524024 | 2.92 |
ENSMUST00000079085.11
|
Rpl34
|
ribosomal protein L34 |
| chrX_+_139857688 | 2.91 |
ENSMUST00000239541.1
|
Atg4a
|
autophagy related 4A, cysteine peptidase |
| chr14_+_62529924 | 2.83 |
ENSMUST00000166879.8
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chr4_-_131802606 | 2.79 |
ENSMUST00000146021.8
|
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr10_-_129738595 | 2.75 |
ENSMUST00000071557.2
|
Olfr815
|
olfactory receptor 815 |
| chr2_+_91376650 | 2.73 |
ENSMUST00000099716.11
ENSMUST00000046769.16 ENSMUST00000111337.3 |
Ckap5
|
cytoskeleton associated protein 5 |
| chr12_+_108572015 | 2.72 |
ENSMUST00000109854.9
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chr19_-_6065872 | 2.69 |
ENSMUST00000164843.10
|
Capn1
|
calpain 1 |
| chr19_+_53128901 | 2.69 |
ENSMUST00000235754.2
ENSMUST00000237301.2 ENSMUST00000238130.2 |
Add3
|
adducin 3 (gamma) |
| chr9_+_118892497 | 2.63 |
ENSMUST00000141185.8
ENSMUST00000126251.8 ENSMUST00000136561.2 |
Vill
|
villin-like |
| chr6_-_115014777 | 2.54 |
ENSMUST00000174848.8
ENSMUST00000032461.12 |
Tamm41
|
TAM41 mitochondrial translocator assembly and maintenance homolog |
| chr17_-_31348576 | 2.52 |
ENSMUST00000024827.5
|
Tff3
|
trefoil factor 3, intestinal |
| chr1_+_40478926 | 2.45 |
ENSMUST00000173514.8
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr1_+_107456731 | 2.44 |
ENSMUST00000182198.8
|
Serpinb10
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr11_-_113574981 | 2.43 |
ENSMUST00000120194.2
|
Fam104a
|
family with sequence similarity 104, member A |
| chr11_-_69786324 | 2.33 |
ENSMUST00000001631.7
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr10_-_62258195 | 2.31 |
ENSMUST00000020277.9
|
Hkdc1
|
hexokinase domain containing 1 |
| chr19_+_34078333 | 2.28 |
ENSMUST00000025685.8
|
Lipm
|
lipase, family member M |
| chr4_-_155012643 | 2.25 |
ENSMUST00000123514.8
|
Tnfrsf14
|
tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) |
| chr2_+_36120438 | 2.22 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr16_+_38167352 | 2.20 |
ENSMUST00000050273.9
ENSMUST00000120495.2 ENSMUST00000119704.2 |
Cox17
Gm21987
|
cytochrome c oxidase assembly protein 17, copper chaperone predicted gene 21987 |
| chr6_-_122317484 | 2.13 |
ENSMUST00000112600.9
|
Phc1
|
polyhomeotic 1 |
| chr3_-_86827640 | 2.06 |
ENSMUST00000195561.6
|
Dclk2
|
doublecortin-like kinase 2 |
| chr1_+_82817794 | 2.04 |
ENSMUST00000186043.2
|
Agfg1
|
ArfGAP with FG repeats 1 |
| chr9_+_21437440 | 2.04 |
ENSMUST00000086361.12
ENSMUST00000173769.3 |
AB124611
|
cDNA sequence AB124611 |
| chr7_-_24705320 | 2.04 |
ENSMUST00000102858.10
ENSMUST00000196684.2 ENSMUST00000080882.11 |
Atp1a3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr8_-_85567256 | 2.03 |
ENSMUST00000003911.13
ENSMUST00000109761.9 ENSMUST00000128035.2 |
Rad23a
|
RAD23 homolog A, nucleotide excision repair protein |
| chrX_+_133486391 | 2.03 |
ENSMUST00000113211.8
|
Rpl36a
|
ribosomal protein L36A |
| chr8_+_23901506 | 1.97 |
ENSMUST00000033952.8
|
Sfrp1
|
secreted frizzled-related protein 1 |
| chr3_-_86827664 | 1.92 |
ENSMUST00000194452.2
ENSMUST00000191752.6 |
Dclk2
|
doublecortin-like kinase 2 |
| chr15_-_99149810 | 1.91 |
ENSMUST00000163506.3
ENSMUST00000229671.2 ENSMUST00000229359.2 ENSMUST00000041190.17 |
Mcrs1
|
microspherule protein 1 |
| chr19_-_34143437 | 1.87 |
ENSMUST00000025686.9
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr1_+_160898283 | 1.87 |
ENSMUST00000028035.14
ENSMUST00000111620.10 ENSMUST00000111618.8 |
Cenpl
|
centromere protein L |
| chr19_-_4240984 | 1.86 |
ENSMUST00000045864.4
|
Tbc1d10c
|
TBC1 domain family, member 10c |
| chr3_-_72875187 | 1.85 |
ENSMUST00000167334.8
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr8_-_4829473 | 1.83 |
ENSMUST00000207262.2
|
Shcbp1
|
Shc SH2-domain binding protein 1 |
| chr2_-_85966272 | 1.81 |
ENSMUST00000216566.3
ENSMUST00000214364.2 |
Olfr1039
|
olfactory receptor 1039 |
| chr17_+_35133435 | 1.80 |
ENSMUST00000007249.15
|
Slc44a4
|
solute carrier family 44, member 4 |
| chr5_+_136023649 | 1.77 |
ENSMUST00000111142.9
ENSMUST00000111145.10 ENSMUST00000111144.8 ENSMUST00000199239.5 ENSMUST00000005072.10 ENSMUST00000130345.2 |
Dtx2
|
deltex 2, E3 ubiquitin ligase |
| chr2_+_69652714 | 1.77 |
ENSMUST00000053087.4
|
Klhl23
|
kelch-like 23 |
| chr1_+_34511793 | 1.77 |
ENSMUST00000188972.3
|
Ptpn18
|
protein tyrosine phosphatase, non-receptor type 18 |
| chr14_-_52341426 | 1.77 |
ENSMUST00000227536.2
ENSMUST00000227195.2 ENSMUST00000228815.2 ENSMUST00000228198.2 ENSMUST00000227458.2 ENSMUST00000228232.2 ENSMUST00000227242.2 ENSMUST00000228748.2 |
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr10_-_75946790 | 1.76 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr10_-_30076543 | 1.73 |
ENSMUST00000099985.6
|
Cenpw
|
centromere protein W |
| chr14_-_86986541 | 1.73 |
ENSMUST00000226254.2
|
Diaph3
|
diaphanous related formin 3 |
| chr5_-_148988110 | 1.73 |
ENSMUST00000110505.8
|
Hmgb1
|
high mobility group box 1 |
| chr3_-_20329823 | 1.72 |
ENSMUST00000011607.6
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr15_-_57982705 | 1.72 |
ENSMUST00000228783.2
|
Atad2
|
ATPase family, AAA domain containing 2 |
| chr14_-_52341472 | 1.71 |
ENSMUST00000111610.12
ENSMUST00000164655.2 |
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr16_-_18904240 | 1.69 |
ENSMUST00000103746.3
|
Iglv1
|
immunoglobulin lambda variable 1 |
| chrX_+_164953444 | 1.69 |
ENSMUST00000130880.9
ENSMUST00000056410.11 ENSMUST00000096252.10 ENSMUST00000087169.11 |
Gemin8
|
gem nuclear organelle associated protein 8 |
| chr16_+_49620883 | 1.68 |
ENSMUST00000229640.2
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr1_-_172722589 | 1.68 |
ENSMUST00000027824.7
|
Apcs
|
serum amyloid P-component |
| chr9_-_21671571 | 1.66 |
ENSMUST00000217382.2
ENSMUST00000214149.2 ENSMUST00000098942.6 ENSMUST00000216057.2 |
Spc24
|
SPC24, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr5_-_151574620 | 1.66 |
ENSMUST00000038131.10
|
Rfc3
|
replication factor C (activator 1) 3 |
| chr7_-_103778992 | 1.65 |
ENSMUST00000053743.6
|
Ubqln5
|
ubiquilin 5 |
| chr12_-_55061117 | 1.65 |
ENSMUST00000172875.8
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr19_+_53128861 | 1.64 |
ENSMUST00000111741.10
|
Add3
|
adducin 3 (gamma) |
| chr2_-_113678999 | 1.61 |
ENSMUST00000102545.8
ENSMUST00000110948.8 |
Arhgap11a
|
Rho GTPase activating protein 11A |
| chr3_-_88317601 | 1.61 |
ENSMUST00000193338.6
ENSMUST00000056370.13 |
Pmf1
|
polyamine-modulated factor 1 |
| chr17_-_35246886 | 1.59 |
ENSMUST00000040151.9
|
Sapcd1
|
suppressor APC domain containing 1 |
| chr14_+_32043944 | 1.57 |
ENSMUST00000022480.8
ENSMUST00000228529.2 |
Ogdhl
|
oxoglutarate dehydrogenase-like |
| chr15_-_100322089 | 1.56 |
ENSMUST00000154331.2
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr9_+_113641615 | 1.56 |
ENSMUST00000111838.10
ENSMUST00000166734.10 ENSMUST00000214522.2 ENSMUST00000163895.3 |
Clasp2
|
CLIP associating protein 2 |
| chr6_+_30541581 | 1.53 |
ENSMUST00000096066.5
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr15_-_100322934 | 1.52 |
ENSMUST00000123461.8
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr13_+_51799268 | 1.51 |
ENSMUST00000075853.6
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr14_+_79753055 | 1.51 |
ENSMUST00000110835.3
ENSMUST00000227192.2 |
Elf1
|
E74-like factor 1 |
| chr5_-_137856280 | 1.51 |
ENSMUST00000110978.7
ENSMUST00000199387.2 ENSMUST00000196195.2 |
Pilrb1
|
paired immunoglobin-like type 2 receptor beta 1 |
| chr4_+_34893772 | 1.50 |
ENSMUST00000029975.10
ENSMUST00000135871.8 ENSMUST00000108130.2 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr2_+_87610895 | 1.50 |
ENSMUST00000215394.2
|
Olfr152
|
olfactory receptor 152 |
| chr13_-_95661726 | 1.49 |
ENSMUST00000022185.10
|
F2rl1
|
coagulation factor II (thrombin) receptor-like 1 |
| chr11_-_78074377 | 1.49 |
ENSMUST00000102483.5
|
Rpl23a
|
ribosomal protein L23A |
| chr8_-_62576140 | 1.48 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr5_-_137530214 | 1.47 |
ENSMUST00000140139.2
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr19_+_13208692 | 1.45 |
ENSMUST00000207246.4
|
Olfr1463
|
olfactory receptor 1463 |
| chr11_-_73382303 | 1.42 |
ENSMUST00000119863.2
ENSMUST00000215358.2 ENSMUST00000214623.2 |
Olfr381
|
olfactory receptor 381 |
| chr2_+_91480460 | 1.41 |
ENSMUST00000111331.9
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr2_+_163500290 | 1.40 |
ENSMUST00000164399.8
ENSMUST00000064703.13 ENSMUST00000099105.9 ENSMUST00000152418.8 ENSMUST00000126182.8 ENSMUST00000131228.8 |
Pkig
|
protein kinase inhibitor, gamma |
| chr13_-_23735822 | 1.39 |
ENSMUST00000102971.2
|
H4c6
|
H4 clustered histone 6 |
| chr15_-_63869818 | 1.38 |
ENSMUST00000164532.3
|
Cyrib
|
CYFIP related Rac1 interactor B |
| chr5_-_44139099 | 1.38 |
ENSMUST00000061299.9
|
Fgfbp1
|
fibroblast growth factor binding protein 1 |
| chr19_+_46385321 | 1.38 |
ENSMUST00000039922.13
ENSMUST00000111867.9 ENSMUST00000120778.8 |
Sufu
|
SUFU negative regulator of hedgehog signaling |
| chr6_-_69282389 | 1.37 |
ENSMUST00000103350.3
|
Igkv4-68
|
immunoglobulin kappa variable 4-68 |
| chr6_+_41331039 | 1.37 |
ENSMUST00000072103.7
|
Try10
|
trypsin 10 |
| chr19_-_11582207 | 1.33 |
ENSMUST00000025582.11
|
Ms4a6d
|
membrane-spanning 4-domains, subfamily A, member 6D |
| chr19_+_13339600 | 1.33 |
ENSMUST00000215096.2
|
Olfr1467
|
olfactory receptor 1467 |
| chr3_+_88486908 | 1.33 |
ENSMUST00000195014.6
|
Ssr2
|
signal sequence receptor, beta |
| chr5_-_18093739 | 1.32 |
ENSMUST00000169095.6
ENSMUST00000197574.2 |
Cd36
|
CD36 molecule |
| chr6_-_68713748 | 1.31 |
ENSMUST00000183936.2
ENSMUST00000196863.2 |
Igkv19-93
|
immunoglobulin kappa chain variable 19-93 |
| chr18_+_44237474 | 1.30 |
ENSMUST00000081271.7
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr2_-_119985078 | 1.29 |
ENSMUST00000028755.8
|
Ehd4
|
EH-domain containing 4 |
| chr5_-_137834470 | 1.29 |
ENSMUST00000110980.2
ENSMUST00000058897.11 ENSMUST00000199028.2 |
Pilra
|
paired immunoglobin-like type 2 receptor alpha |
| chr14_-_104760051 | 1.27 |
ENSMUST00000022716.4
ENSMUST00000228448.2 ENSMUST00000227640.2 |
Obi1
|
ORC ubiquitin ligase 1 |
| chr14_+_61547202 | 1.27 |
ENSMUST00000055159.8
|
Arl11
|
ADP-ribosylation factor-like 11 |
| chr6_+_136509922 | 1.26 |
ENSMUST00000187429.4
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr9_-_123507937 | 1.25 |
ENSMUST00000040960.13
|
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr8_+_66838927 | 1.24 |
ENSMUST00000039540.12
ENSMUST00000110253.3 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr15_-_99149794 | 1.24 |
ENSMUST00000229926.2
|
Mcrs1
|
microspherule protein 1 |
| chr9_+_19828161 | 1.23 |
ENSMUST00000217347.2
ENSMUST00000057596.10 |
Olfr77
|
olfactory receptor 77 |
| chr18_-_67378886 | 1.22 |
ENSMUST00000073054.5
|
Mppe1
|
metallophosphoesterase 1 |
| chr11_+_87000032 | 1.20 |
ENSMUST00000020794.6
|
Ska2
|
spindle and kinetochore associated complex subunit 2 |
| chr5_+_117378510 | 1.20 |
ENSMUST00000111975.3
|
Taok3
|
TAO kinase 3 |
| chr6_-_87815653 | 1.20 |
ENSMUST00000204431.2
ENSMUST00000089497.7 |
Isy1
|
ISY1 splicing factor homolog |
| chr10_+_58159288 | 1.19 |
ENSMUST00000020078.14
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr2_+_155593030 | 1.19 |
ENSMUST00000029140.12
ENSMUST00000132608.2 |
Procr
|
protein C receptor, endothelial |
| chr5_-_138185438 | 1.17 |
ENSMUST00000110937.8
ENSMUST00000139276.2 ENSMUST00000048698.14 ENSMUST00000123415.8 |
Taf6
|
TATA-box binding protein associated factor 6 |
| chr4_-_41464816 | 1.16 |
ENSMUST00000108055.9
ENSMUST00000154535.8 ENSMUST00000030148.6 |
Kif24
|
kinesin family member 24 |
| chr7_+_97480125 | 1.16 |
ENSMUST00000206351.2
|
Pak1
|
p21 (RAC1) activated kinase 1 |
| chr6_+_47897410 | 1.15 |
ENSMUST00000009411.9
|
Zfp212
|
Zinc finger protein 212 |
| chr3_-_116047148 | 1.15 |
ENSMUST00000090473.7
|
Gpr88
|
G-protein coupled receptor 88 |
| chr17_+_66418525 | 1.15 |
ENSMUST00000072383.14
|
Washc1
|
WASH complex subunit 1 |
| chr2_-_144112700 | 1.14 |
ENSMUST00000110030.10
|
Snx5
|
sorting nexin 5 |
| chr2_+_91480513 | 1.13 |
ENSMUST00000090614.11
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr19_-_6065799 | 1.13 |
ENSMUST00000235138.2
|
Capn1
|
calpain 1 |
| chr5_-_108943211 | 1.12 |
ENSMUST00000004943.2
|
Tmed11
|
transmembrane p24 trafficking protein 11 |
| chr4_+_19818718 | 1.11 |
ENSMUST00000035890.8
|
Slc7a13
|
solute carrier family 7, (cationic amino acid transporter, y+ system) member 13 |
| chr7_+_45271229 | 1.11 |
ENSMUST00000033100.5
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr3_-_151899470 | 1.10 |
ENSMUST00000050073.13
|
Dnajb4
|
DnaJ heat shock protein family (Hsp40) member B4 |
| chr1_-_126758369 | 1.10 |
ENSMUST00000112583.8
ENSMUST00000094609.10 |
Nckap5
|
NCK-associated protein 5 |
| chr3_-_75177378 | 1.10 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr3_-_113325938 | 1.09 |
ENSMUST00000132353.2
|
Amy2a1
|
amylase 2a1 |
| chr6_-_29212239 | 1.09 |
ENSMUST00000160878.8
|
Impdh1
|
inosine monophosphate dehydrogenase 1 |
| chr5_-_44139121 | 1.08 |
ENSMUST00000199894.2
|
Fgfbp1
|
fibroblast growth factor binding protein 1 |
| chr3_+_88486921 | 1.08 |
ENSMUST00000193934.6
ENSMUST00000192495.6 ENSMUST00000035785.9 |
Ssr2
|
signal sequence receptor, beta |
| chr18_-_32044877 | 1.08 |
ENSMUST00000054984.8
|
Sft2d3
|
SFT2 domain containing 3 |
| chr10_+_87041814 | 1.08 |
ENSMUST00000189775.2
|
1700113H08Rik
|
RIKEN cDNA 1700113H08 gene |
| chr1_+_139382485 | 1.08 |
ENSMUST00000200083.5
ENSMUST00000053364.12 |
Aspm
|
abnormal spindle microtubule assembly |
| chr9_-_123507847 | 1.07 |
ENSMUST00000170591.2
ENSMUST00000171647.9 |
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr19_-_32173824 | 1.07 |
ENSMUST00000151822.2
|
Sgms1
|
sphingomyelin synthase 1 |
| chr4_+_134658209 | 1.07 |
ENSMUST00000030622.3
|
Syf2
|
SYF2 homolog, RNA splicing factor (S. cerevisiae) |
| chr5_-_65855511 | 1.06 |
ENSMUST00000201948.4
|
Pds5a
|
PDS5 cohesin associated factor A |
| chr18_-_43610829 | 1.04 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr1_-_134883645 | 1.04 |
ENSMUST00000045665.13
ENSMUST00000086444.6 ENSMUST00000112163.2 |
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr7_-_43309563 | 1.04 |
ENSMUST00000032667.10
|
Siglece
|
sialic acid binding Ig-like lectin E |
| chr5_-_137531471 | 1.02 |
ENSMUST00000143495.8
ENSMUST00000111020.8 ENSMUST00000111023.8 ENSMUST00000111038.8 |
Gnb2
Epo
|
guanine nucleotide binding protein (G protein), beta 2 erythropoietin |
| chr6_+_135339929 | 1.02 |
ENSMUST00000032330.16
|
Emp1
|
epithelial membrane protein 1 |
| chr11_+_113575208 | 1.02 |
ENSMUST00000042227.15
ENSMUST00000123466.2 ENSMUST00000106621.4 |
D11Wsu47e
|
DNA segment, Chr 11, Wayne State University 47, expressed |
| chr7_+_108265625 | 1.00 |
ENSMUST00000213979.3
ENSMUST00000216331.2 ENSMUST00000217170.2 |
Olfr510
|
olfactory receptor 510 |
| chr2_-_126342551 | 1.00 |
ENSMUST00000129187.2
|
Atp8b4
|
ATPase, class I, type 8B, member 4 |
| chr11_+_46701619 | 1.00 |
ENSMUST00000068877.7
|
Timd4
|
T cell immunoglobulin and mucin domain containing 4 |
| chr7_-_100232276 | 0.99 |
ENSMUST00000152876.3
ENSMUST00000150042.8 ENSMUST00000132888.9 |
Mrpl48
|
mitochondrial ribosomal protein L48 |
| chr4_+_59035088 | 0.98 |
ENSMUST00000041160.13
|
Gng10
|
guanine nucleotide binding protein (G protein), gamma 10 |
| chr7_-_103094646 | 0.98 |
ENSMUST00000215417.2
|
Olfr605
|
olfactory receptor 605 |
| chr18_+_23886765 | 0.98 |
ENSMUST00000115830.8
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr2_+_29236815 | 0.97 |
ENSMUST00000028139.11
ENSMUST00000113830.11 |
Med27
|
mediator complex subunit 27 |
| chr3_-_19319155 | 0.97 |
ENSMUST00000091314.11
|
Pde7a
|
phosphodiesterase 7A |
| chrX_+_158623460 | 0.97 |
ENSMUST00000112451.8
ENSMUST00000112453.9 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr19_-_38032006 | 0.97 |
ENSMUST00000172095.3
ENSMUST00000041475.16 |
Myof
|
myoferlin |
| chr9_+_19716202 | 0.95 |
ENSMUST00000212540.3
ENSMUST00000217280.2 |
Olfr859
|
olfactory receptor 859 |
| chr3_+_66127330 | 0.94 |
ENSMUST00000029421.6
|
Ptx3
|
pentraxin related gene |
| chr15_-_81614063 | 0.94 |
ENSMUST00000171115.3
ENSMUST00000170134.9 ENSMUST00000052374.13 |
Rangap1
|
RAN GTPase activating protein 1 |
| chr8_+_107757847 | 0.94 |
ENSMUST00000034388.10
|
Vps4a
|
vacuolar protein sorting 4A |
| chr15_-_100321973 | 0.94 |
ENSMUST00000154676.2
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr19_+_12647803 | 0.94 |
ENSMUST00000207341.3
ENSMUST00000208494.3 ENSMUST00000208657.3 |
Olfr1442
|
olfactory receptor 1442 |
| chr5_-_137531463 | 0.94 |
ENSMUST00000170293.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr11_-_115918784 | 0.93 |
ENSMUST00000106454.8
|
H3f3b
|
H3.3 histone B |
| chr7_-_30523191 | 0.92 |
ENSMUST00000053156.10
|
Ffar2
|
free fatty acid receptor 2 |
| chr1_+_134333720 | 0.91 |
ENSMUST00000173908.8
|
Cyb5r1
|
cytochrome b5 reductase 1 |
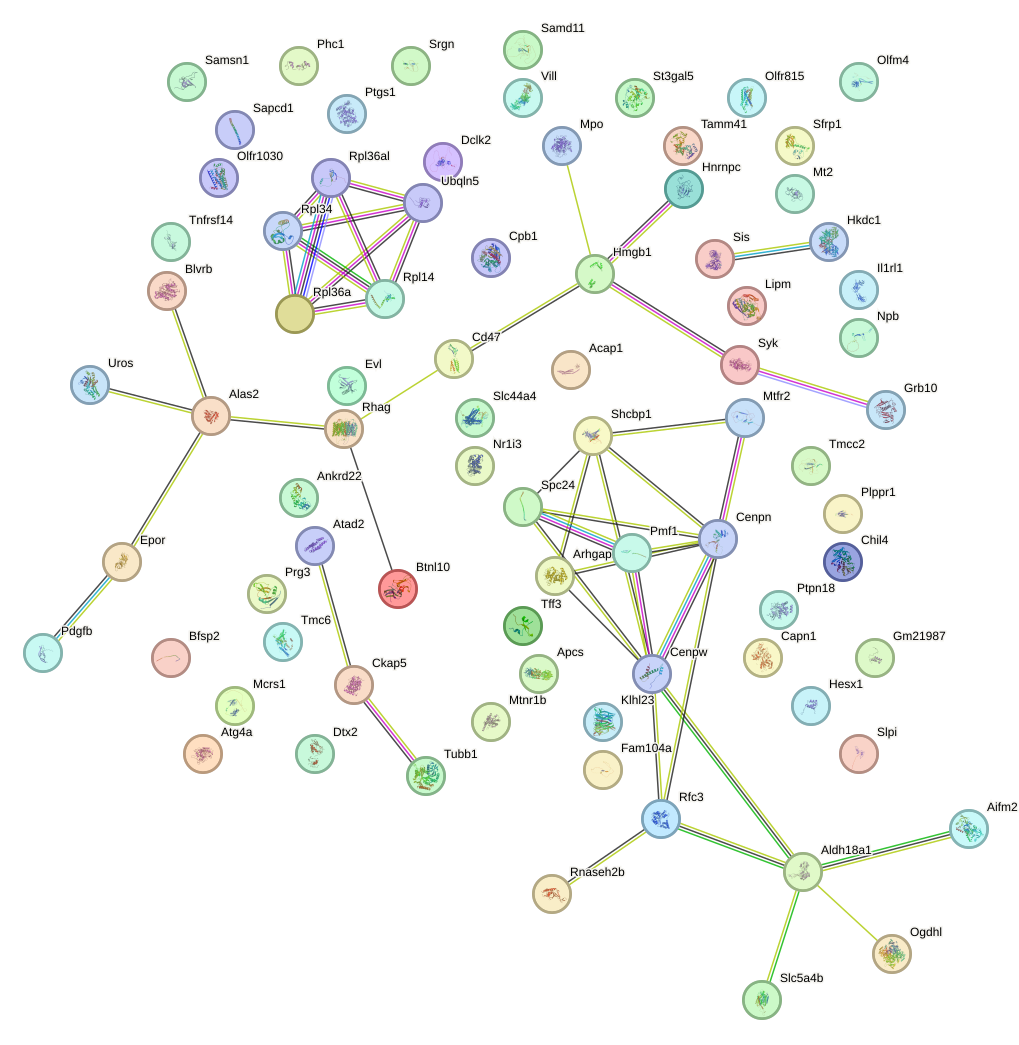
Network of associatons between targets according to the STRING database.
First level regulatory network of Dlx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 19.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 4.0 | 12.1 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 3.1 | 9.2 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 2.1 | 6.2 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 1.9 | 7.7 | GO:0045575 | basophil activation(GO:0045575) |
| 1.2 | 3.5 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 1.1 | 4.4 | GO:0033375 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 1.1 | 9.8 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.9 | 6.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.9 | 2.6 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.8 | 4.9 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.8 | 3.0 | GO:0045425 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) cellular response to molecule of fungal origin(GO:0071226) |
| 0.7 | 2.2 | GO:1904959 | regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.7 | 4.0 | GO:0015692 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.7 | 6.6 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.7 | 2.0 | GO:2000040 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.6 | 2.5 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.6 | 1.8 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.6 | 2.3 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.6 | 1.7 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.6 | 1.7 | GO:0002545 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.5 | 9.9 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.5 | 3.2 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.5 | 6.2 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.5 | 1.5 | GO:1900135 | positive regulation of eosinophil degranulation(GO:0043311) regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) positive regulation of renin secretion into blood stream(GO:1900135) positive regulation of eosinophil activation(GO:1902568) |
| 0.5 | 2.7 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.4 | 3.6 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.4 | 1.3 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.4 | 0.8 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.4 | 1.2 | GO:0000389 | generation of catalytic spliceosome for second transesterification step(GO:0000350) mRNA 3'-splice site recognition(GO:0000389) |
| 0.4 | 3.1 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.3 | 4.9 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.3 | 5.9 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.3 | 2.2 | GO:0035633 | cyclooxygenase pathway(GO:0019371) maintenance of blood-brain barrier(GO:0035633) |
| 0.3 | 0.9 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.3 | 3.4 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.3 | 1.2 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.3 | 1.3 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.3 | 1.0 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 0.2 | 2.5 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.2 | 2.5 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.2 | 0.9 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.2 | 0.8 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.2 | 1.7 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.2 | 1.9 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.2 | 3.1 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.2 | 3.8 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.2 | 1.4 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.2 | 1.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.2 | 0.8 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) meiotic metaphase plate congression(GO:0051311) |
| 0.2 | 1.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 0.9 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.2 | 2.7 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.2 | 0.7 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.2 | 3.9 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.2 | 1.7 | GO:0008228 | opsonization(GO:0008228) |
| 0.2 | 0.5 | GO:0032685 | negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) |
| 0.2 | 2.7 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.2 | 1.4 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.2 | 1.6 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 | 1.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.2 | 4.9 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.1 | 0.7 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.1 | 0.7 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.1 | 0.7 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 1.5 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 | 0.8 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 2.0 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 0.1 | 9.8 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.9 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 | 0.7 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 2.8 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.3 | GO:0060003 | copper ion export(GO:0060003) |
| 0.1 | 1.8 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.5 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 8.4 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.1 | 1.7 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.4 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.1 | 9.1 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.1 | 0.5 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.1 | 2.4 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 | 0.3 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.1 | 0.1 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 0.8 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.1 | 0.9 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.1 | 1.7 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.4 | GO:1904882 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.1 | 0.7 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 2.0 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 2.3 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.1 | 0.8 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 2.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 0.2 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.1 | 0.6 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.1 | 1.5 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 0.2 | GO:0060448 | epicardial cell to mesenchymal cell transition(GO:0003347) dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.1 | 0.6 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.1 | 4.6 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 1.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.6 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 | 1.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.3 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 3.3 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 5.6 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 0.4 | GO:0044334 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.1 | 2.8 | GO:0002820 | negative regulation of adaptive immune response(GO:0002820) |
| 0.1 | 1.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 | 0.3 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.1 | 0.7 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.9 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.2 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.1 | 0.7 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.1 | 0.8 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 6.0 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 1.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.5 | GO:0002441 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 | 0.2 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.6 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.2 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.1 | 0.3 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 1.7 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 0.2 | GO:0060382 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 0.1 | 1.6 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 0.3 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.1 | 0.7 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 0.3 | GO:1903207 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.1 | 0.3 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.1 | 1.7 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.1 | 2.5 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 1.2 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 | 0.4 | GO:0038109 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Fc-epsilon receptor signaling pathway(GO:0038095) Kit signaling pathway(GO:0038109) |
| 0.1 | 0.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.4 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 3.5 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.4 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 1.3 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.5 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.8 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.8 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 8.2 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.6 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.3 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.2 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 2.2 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 32.8 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 1.7 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 1.1 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.7 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.3 | GO:0071231 | neural crest cell migration involved in heart formation(GO:0003147) folic acid metabolic process(GO:0046655) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.0 | 2.1 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 2.6 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.5 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 3.7 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 2.7 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.7 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 1.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 7.0 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.3 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.0 | 3.1 | GO:2000177 | regulation of neural precursor cell proliferation(GO:2000177) |
| 0.0 | 0.3 | GO:0002579 | positive regulation of antigen processing and presentation(GO:0002579) |
| 0.0 | 0.2 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.0 | 0.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.7 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.0 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.0 | 0.5 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.1 | GO:0072161 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.2 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 2.6 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.7 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.1 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.0 | 0.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.3 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 1.9 | GO:0031109 | microtubule polymerization or depolymerization(GO:0031109) |
| 0.0 | 1.5 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 3.1 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.3 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.2 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 1.3 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.1 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.0 | 0.0 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:0042118 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) endothelial cell activation(GO:0042118) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 7.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.7 | 12.1 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.7 | 4.0 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.5 | 9.5 | GO:0042581 | specific granule(GO:0042581) |
| 0.5 | 1.6 | GO:1903754 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.5 | 2.8 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.4 | 1.6 | GO:0008623 | CHRAC(GO:0008623) |
| 0.4 | 4.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.3 | 1.6 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.3 | 1.7 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.3 | 1.7 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.3 | 2.4 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.3 | 0.8 | GO:0005712 | chiasma(GO:0005712) late recombination nodule(GO:0005715) |
| 0.2 | 1.7 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.2 | 0.9 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.2 | 0.7 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.2 | 0.6 | GO:0090537 | CERF complex(GO:0090537) |
| 0.2 | 2.4 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.2 | 2.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 0.9 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.2 | 0.8 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.2 | 3.9 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 0.7 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 1.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 3.2 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 1.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) tubular endosome(GO:0097422) |
| 0.1 | 0.4 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 10.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.5 | GO:0071920 | cleavage body(GO:0071920) |
| 0.1 | 1.7 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 3.5 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 0.4 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 1.6 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.3 | GO:0042025 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.1 | 5.6 | GO:1990752 | microtubule end(GO:1990752) |
| 0.1 | 6.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.6 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 5.5 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 1.9 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 0.9 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 1.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.9 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.8 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 1.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.8 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.8 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 1.0 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.3 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.0 | 0.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 1.5 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.4 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.6 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 4.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.6 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.6 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 4.9 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 1.0 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 1.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 7.5 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 9.8 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.6 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 11.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.8 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 1.3 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 1.8 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 3.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.4 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 1.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.3 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 8.7 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 1.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.4 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 4.4 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 17.2 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 2.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.8 | GO:0000502 | proteasome complex(GO:0000502) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.2 | GO:0017084 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 3.0 | 8.9 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 2.1 | 6.3 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 1.2 | 3.6 | GO:0042602 | riboflavin reductase (NADPH) activity(GO:0042602) |
| 1.1 | 9.8 | GO:0004568 | chitinase activity(GO:0004568) |
| 1.0 | 3.1 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 1.0 | 6.2 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.9 | 3.7 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.9 | 19.1 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.8 | 2.5 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.8 | 0.8 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.7 | 4.0 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.6 | 1.7 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.5 | 1.9 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.4 | 2.8 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.4 | 3.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.4 | 1.5 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.3 | 3.6 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.3 | 2.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.3 | 1.3 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.3 | 3.0 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.3 | 2.3 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.3 | 3.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.3 | 1.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.3 | 1.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 0.7 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.2 | 1.6 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.2 | 1.8 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.2 | 1.5 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 1.4 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 14.3 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.2 | 0.8 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.2 | 1.8 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.2 | 1.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.2 | 4.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 0.5 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.2 | 2.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 1.0 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.2 | 1.6 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 1.4 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.2 | 1.7 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.7 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 2.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.8 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 3.0 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.1 | 1.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 1.9 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 4.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.3 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.1 | 0.7 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 3.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.5 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.5 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 3.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 9.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.7 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 3.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.6 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 2.0 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 0.5 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.1 | 0.4 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.1 | 1.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 7.9 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 4.9 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 1.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.7 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 1.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.4 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.1 | 0.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.4 | GO:0032405 | MutLalpha complex binding(GO:0032405) |
| 0.1 | 0.7 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 23.6 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 0.9 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 1.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 0.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 4.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.5 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 3.1 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 6.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.4 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.1 | 1.0 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.3 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 0.3 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.1 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 1.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.3 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.5 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.2 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 1.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 2.0 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 1.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.3 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.2 | GO:0016623 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 6.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 4.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 6.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 18.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.3 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 1.0 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 3.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 1.2 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 1.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 5.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.8 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 1.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.7 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.8 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 3.4 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 2.9 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.2 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) |
| 0.0 | 0.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.8 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 1.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 2.1 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 1.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 2.4 | GO:0008017 | microtubule binding(GO:0008017) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.8 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.3 | 12.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 10.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 12.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 5.9 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 3.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 1.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 3.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 3.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 4.0 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 1.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 3.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 2.5 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.9 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.6 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.8 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 2.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.9 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 2.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 2.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.9 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.9 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 1.3 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 3.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 18.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.5 | 20.4 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.3 | 6.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.3 | 9.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 6.6 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 8.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 6.3 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.2 | 1.5 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 3.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.7 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.7 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.0 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 3.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 10.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.7 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 2.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 0.7 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 9.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 0.8 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 2.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 3.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 2.4 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.1 | 0.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.7 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.1 | 2.7 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.1 | 1.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 2.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 2.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 2.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 5.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.8 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 2.2 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.8 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.8 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 5.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.6 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.8 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 2.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 2.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |