Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Dmc1
Z-value: 0.82
Transcription factors associated with Dmc1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Dmc1
|
ENSMUSG00000022429.12 | DNA meiotic recombinase 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dmc1 | mm39_v1_chr15_-_79489286_79489315 | -0.31 | 7.0e-02 | Click! |
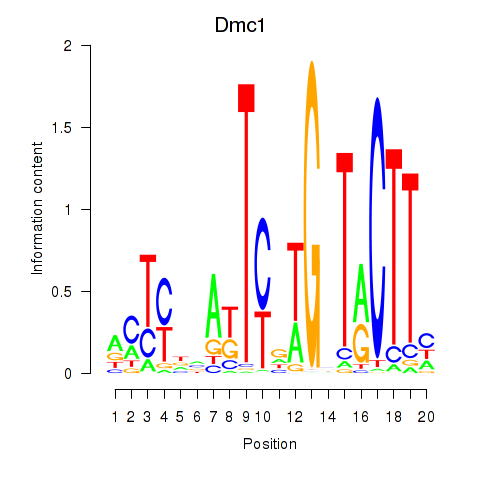
Activity profile of Dmc1 motif
Sorted Z-values of Dmc1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_87485023 | 4.47 |
ENSMUST00000031195.3
|
Ugt2a3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr19_+_40078132 | 4.20 |
ENSMUST00000068094.13
ENSMUST00000080171.3 |
Cyp2c50
|
cytochrome P450, family 2, subfamily c, polypeptide 50 |
| chr4_-_96552349 | 3.23 |
ENSMUST00000030299.8
|
Cyp2j5
|
cytochrome P450, family 2, subfamily j, polypeptide 5 |
| chr5_-_87074380 | 3.06 |
ENSMUST00000031183.3
|
Ugt2b1
|
UDP glucuronosyltransferase 2 family, polypeptide B1 |
| chr13_-_56696310 | 2.98 |
ENSMUST00000062806.6
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr13_-_56696222 | 2.76 |
ENSMUST00000225183.2
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr4_-_107164315 | 2.63 |
ENSMUST00000126291.2
ENSMUST00000106748.2 ENSMUST00000129138.2 |
Dio1
|
deiodinase, iodothyronine, type I |
| chr5_-_87054796 | 2.32 |
ENSMUST00000031181.16
ENSMUST00000113333.2 |
Ugt2b34
|
UDP glucuronosyltransferase 2 family, polypeptide B34 |
| chr7_-_12732067 | 2.31 |
ENSMUST00000032539.14
ENSMUST00000120903.8 |
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr7_-_12731594 | 2.27 |
ENSMUST00000133977.3
|
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr19_-_40062174 | 2.02 |
ENSMUST00000048959.5
|
Cyp2c54
|
cytochrome P450, family 2, subfamily c, polypeptide 54 |
| chr19_-_46661321 | 2.00 |
ENSMUST00000026012.8
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr19_-_7779943 | 2.00 |
ENSMUST00000120522.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr17_-_33136021 | 1.96 |
ENSMUST00000054174.9
|
Cyp4f14
|
cytochrome P450, family 4, subfamily f, polypeptide 14 |
| chr10_+_87697155 | 1.95 |
ENSMUST00000122100.3
|
Igf1
|
insulin-like growth factor 1 |
| chr4_-_107928567 | 1.92 |
ENSMUST00000106701.2
|
Scp2
|
sterol carrier protein 2, liver |
| chr19_-_7780025 | 1.85 |
ENSMUST00000065634.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr13_-_24098981 | 1.80 |
ENSMUST00000110407.4
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr10_+_127637015 | 1.78 |
ENSMUST00000071646.2
|
Rdh16
|
retinol dehydrogenase 16 |
| chr14_+_30608433 | 1.77 |
ENSMUST00000120269.11
ENSMUST00000078490.14 ENSMUST00000006703.15 |
Itih4
|
inter alpha-trypsin inhibitor, heavy chain 4 |
| chr14_+_30608478 | 1.77 |
ENSMUST00000168782.4
|
Itih4
|
inter alpha-trypsin inhibitor, heavy chain 4 |
| chr6_+_121983720 | 1.66 |
ENSMUST00000081777.8
|
Mug2
|
murinoglobulin 2 |
| chr8_+_105460627 | 1.65 |
ENSMUST00000034346.15
ENSMUST00000164182.3 |
Ces2a
|
carboxylesterase 2A |
| chr1_-_192946359 | 1.54 |
ENSMUST00000161737.8
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr5_+_87148697 | 1.51 |
ENSMUST00000031186.9
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr19_+_39980868 | 1.49 |
ENSMUST00000049178.3
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr13_-_24098951 | 1.42 |
ENSMUST00000021769.16
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr7_-_27037096 | 1.33 |
ENSMUST00000038618.13
ENSMUST00000108369.9 |
Ltbp4
|
latent transforming growth factor beta binding protein 4 |
| chr3_-_63836796 | 1.30 |
ENSMUST00000061706.7
|
E130311K13Rik
|
RIKEN cDNA E130311K13 gene |
| chr5_-_89583469 | 1.28 |
ENSMUST00000200534.2
|
Gc
|
vitamin D binding protein |
| chr5_-_87572060 | 1.25 |
ENSMUST00000072818.6
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr19_-_40175709 | 1.25 |
ENSMUST00000051846.13
|
Cyp2c70
|
cytochrome P450, family 2, subfamily c, polypeptide 70 |
| chr4_+_133280680 | 1.24 |
ENSMUST00000042706.3
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr19_-_46661501 | 1.19 |
ENSMUST00000236174.2
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr17_-_57535003 | 1.19 |
ENSMUST00000177046.2
ENSMUST00000024988.15 |
C3
|
complement component 3 |
| chr17_+_64907697 | 1.15 |
ENSMUST00000086723.10
|
Man2a1
|
mannosidase 2, alpha 1 |
| chr1_+_88093726 | 1.14 |
ENSMUST00000097659.5
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr18_-_56705960 | 1.14 |
ENSMUST00000174518.8
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr3_+_94284739 | 1.11 |
ENSMUST00000197040.5
|
Rorc
|
RAR-related orphan receptor gamma |
| chr19_-_8196196 | 1.10 |
ENSMUST00000113298.9
|
Slc22a29
|
solute carrier family 22. member 29 |
| chr3_-_73615732 | 1.03 |
ENSMUST00000029367.6
|
Bche
|
butyrylcholinesterase |
| chr9_+_92157655 | 0.96 |
ENSMUST00000034932.14
ENSMUST00000180154.8 |
Plscr2
|
phospholipid scramblase 2 |
| chr5_-_104169785 | 0.96 |
ENSMUST00000031251.16
|
Hsd17b11
|
hydroxysteroid (17-beta) dehydrogenase 11 |
| chr3_+_20011251 | 0.95 |
ENSMUST00000108328.8
|
Cp
|
ceruloplasmin |
| chr6_-_128503666 | 0.94 |
ENSMUST00000143664.2
ENSMUST00000112132.8 |
Pzp
|
PZP, alpha-2-macroglobulin like |
| chr16_-_56984137 | 0.93 |
ENSMUST00000231733.2
|
Nit2
|
nitrilase family, member 2 |
| chr13_-_92491451 | 0.92 |
ENSMUST00000187424.2
ENSMUST00000185852.7 ENSMUST00000022220.13 |
Msh3
|
mutS homolog 3 |
| chr3_+_94284812 | 0.91 |
ENSMUST00000200009.2
|
Rorc
|
RAR-related orphan receptor gamma |
| chr3_+_20011201 | 0.88 |
ENSMUST00000091309.12
ENSMUST00000108329.8 ENSMUST00000003714.13 |
Cp
|
ceruloplasmin |
| chr4_+_19575128 | 0.87 |
ENSMUST00000108253.8
ENSMUST00000029888.4 |
Rmdn1
|
regulator of microtubule dynamics 1 |
| chr1_+_58152295 | 0.87 |
ENSMUST00000040999.14
ENSMUST00000162011.3 |
Aox3
|
aldehyde oxidase 3 |
| chr1_-_72323464 | 0.86 |
ENSMUST00000027381.13
|
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr5_+_137568086 | 0.86 |
ENSMUST00000198866.5
|
Tfr2
|
transferrin receptor 2 |
| chr1_+_21310821 | 0.86 |
ENSMUST00000121676.8
ENSMUST00000124990.3 |
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr3_+_20011405 | 0.85 |
ENSMUST00000108325.9
|
Cp
|
ceruloplasmin |
| chr4_-_49549489 | 0.84 |
ENSMUST00000029987.10
|
Aldob
|
aldolase B, fructose-bisphosphate |
| chr9_+_111268131 | 0.84 |
ENSMUST00000111879.5
|
Dclk3
|
doublecortin-like kinase 3 |
| chr14_-_61597843 | 0.84 |
ENSMUST00000022494.10
|
Ebpl
|
emopamil binding protein-like |
| chr5_+_90608751 | 0.83 |
ENSMUST00000031314.10
|
Alb
|
albumin |
| chr1_+_21310803 | 0.83 |
ENSMUST00000027067.15
|
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr11_+_48728291 | 0.82 |
ENSMUST00000046903.6
|
Trim7
|
tripartite motif-containing 7 |
| chr1_-_72323407 | 0.80 |
ENSMUST00000097698.5
|
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr13_+_92491234 | 0.80 |
ENSMUST00000022218.6
|
Dhfr
|
dihydrofolate reductase |
| chr2_+_14234198 | 0.80 |
ENSMUST00000028045.4
|
Mrc1
|
mannose receptor, C type 1 |
| chr10_+_105676925 | 0.80 |
ENSMUST00000020049.9
|
Ccdc59
|
coiled-coil domain containing 59 |
| chr11_-_116089866 | 0.80 |
ENSMUST00000066587.12
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr11_-_120618052 | 0.78 |
ENSMUST00000106148.10
ENSMUST00000026144.5 |
Dcxr
|
dicarbonyl L-xylulose reductase |
| chr10_-_128425519 | 0.78 |
ENSMUST00000082059.7
|
Erbb3
|
erb-b2 receptor tyrosine kinase 3 |
| chr4_+_57845240 | 0.78 |
ENSMUST00000102903.8
ENSMUST00000107598.9 |
Pakap
|
paralemmin A kinase anchor protein |
| chr6_+_71176811 | 0.78 |
ENSMUST00000067492.8
|
Fabp1
|
fatty acid binding protein 1, liver |
| chr13_+_30844025 | 0.78 |
ENSMUST00000110310.9
ENSMUST00000095914.7 |
Dusp22
|
dual specificity phosphatase 22 |
| chr9_+_37524966 | 0.78 |
ENSMUST00000215474.2
|
Siae
|
sialic acid acetylesterase |
| chr5_-_87402659 | 0.77 |
ENSMUST00000075858.4
|
Ugt2b37
|
UDP glucuronosyltransferase 2 family, polypeptide B37 |
| chr9_-_58448224 | 0.77 |
ENSMUST00000039788.11
|
Cd276
|
CD276 antigen |
| chr8_+_57004125 | 0.77 |
ENSMUST00000110322.9
ENSMUST00000040218.13 ENSMUST00000210863.2 |
Fbxo8
|
F-box protein 8 |
| chr3_+_135531548 | 0.76 |
ENSMUST00000167390.8
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr2_+_121978156 | 0.75 |
ENSMUST00000102476.5
|
B2m
|
beta-2 microglobulin |
| chr4_+_148686985 | 0.74 |
ENSMUST00000105701.9
ENSMUST00000052060.7 |
Masp2
|
mannan-binding lectin serine peptidase 2 |
| chr6_-_43643093 | 0.74 |
ENSMUST00000114644.8
ENSMUST00000067888.14 |
Tpk1
|
thiamine pyrophosphokinase |
| chr1_+_16735401 | 0.73 |
ENSMUST00000177501.2
ENSMUST00000065373.6 |
Tmem70
|
transmembrane protein 70 |
| chr1_+_131725119 | 0.73 |
ENSMUST00000112393.9
ENSMUST00000048660.12 |
Pm20d1
|
peptidase M20 domain containing 1 |
| chr16_-_10360893 | 0.72 |
ENSMUST00000184863.8
ENSMUST00000038281.6 |
Dexi
|
dexamethasone-induced transcript |
| chr15_-_60696790 | 0.71 |
ENSMUST00000100635.5
|
Lratd2
|
LRAT domain containing 1 |
| chr1_-_183150867 | 0.71 |
ENSMUST00000194543.4
|
Mia3
|
melanoma inhibitory activity 3 |
| chr13_+_33187205 | 0.70 |
ENSMUST00000063191.14
|
Serpinb9
|
serine (or cysteine) peptidase inhibitor, clade B, member 9 |
| chr10_-_95251327 | 0.69 |
ENSMUST00000172070.8
ENSMUST00000150432.8 |
Socs2
|
suppressor of cytokine signaling 2 |
| chr9_+_123921573 | 0.69 |
ENSMUST00000111442.3
ENSMUST00000171499.3 |
Ccr5
|
chemokine (C-C motif) receptor 5 |
| chr9_-_103099262 | 0.69 |
ENSMUST00000170904.2
|
Trf
|
transferrin |
| chr6_-_136852792 | 0.68 |
ENSMUST00000032342.3
|
Mgp
|
matrix Gla protein |
| chr5_+_137568113 | 0.68 |
ENSMUST00000031729.13
ENSMUST00000199054.5 |
Tfr2
|
transferrin receptor 2 |
| chr17_-_74354844 | 0.67 |
ENSMUST00000043458.9
|
Srd5a2
|
steroid 5 alpha-reductase 2 |
| chr3_-_10400710 | 0.67 |
ENSMUST00000078748.4
|
Slc10a5
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 5 |
| chr10_-_89369432 | 0.67 |
ENSMUST00000105297.2
|
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr11_+_99770013 | 0.67 |
ENSMUST00000078442.4
|
Gm11567
|
predicted gene 11567 |
| chr2_+_144369638 | 0.66 |
ENSMUST00000028914.9
ENSMUST00000110017.3 |
Polr3f
|
polymerase (RNA) III (DNA directed) polypeptide F |
| chr4_-_49473904 | 0.66 |
ENSMUST00000135976.2
|
Acnat1
|
acyl-coenzyme A amino acid N-acyltransferase 1 |
| chr14_+_52122439 | 0.66 |
ENSMUST00000167984.2
|
Mettl17
|
methyltransferase like 17 |
| chr11_+_114566257 | 0.65 |
ENSMUST00000045779.6
|
Ttyh2
|
tweety family member 2 |
| chr10_-_95251145 | 0.65 |
ENSMUST00000119917.2
|
Socs2
|
suppressor of cytokine signaling 2 |
| chr8_-_85620537 | 0.65 |
ENSMUST00000003907.14
ENSMUST00000109745.8 ENSMUST00000142748.2 |
Gcdh
|
glutaryl-Coenzyme A dehydrogenase |
| chr1_-_150268771 | 0.65 |
ENSMUST00000097546.9
ENSMUST00000111913.9 |
Odr4
|
odr4 GPCR localization factor homolog |
| chr14_-_76024471 | 0.64 |
ENSMUST00000022580.8
|
Slc25a30
|
solute carrier family 25, member 30 |
| chr16_+_22710134 | 0.63 |
ENSMUST00000231328.2
|
Ahsg
|
alpha-2-HS-glycoprotein |
| chr14_+_52122299 | 0.63 |
ENSMUST00000047899.13
ENSMUST00000164902.8 |
Mettl17
|
methyltransferase like 17 |
| chr4_+_117109148 | 0.63 |
ENSMUST00000062824.12
|
Tmem53
|
transmembrane protein 53 |
| chrM_-_14061 | 0.63 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr4_+_49521176 | 0.62 |
ENSMUST00000042964.13
ENSMUST00000107696.2 |
Zfp189
|
zinc finger protein 189 |
| chrX_-_48377106 | 0.61 |
ENSMUST00000114918.9
ENSMUST00000033437.15 ENSMUST00000114912.8 ENSMUST00000114911.8 |
Enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr4_+_117109204 | 0.61 |
ENSMUST00000125943.8
ENSMUST00000106434.8 |
Tmem53
|
transmembrane protein 53 |
| chr3_+_135531834 | 0.60 |
ENSMUST00000029810.6
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr2_+_113271409 | 0.59 |
ENSMUST00000081349.9
|
Fmn1
|
formin 1 |
| chr7_-_126046814 | 0.58 |
ENSMUST00000146973.2
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr11_+_101442961 | 0.58 |
ENSMUST00000103099.8
|
Nbr1
|
NBR1, autophagy cargo receptor |
| chr15_-_75881289 | 0.57 |
ENSMUST00000170153.2
|
Fam83h
|
family with sequence similarity 83, member H |
| chr7_-_90106375 | 0.56 |
ENSMUST00000032844.7
|
Tmem126a
|
transmembrane protein 126A |
| chr11_+_108271990 | 0.56 |
ENSMUST00000146050.2
ENSMUST00000152958.8 |
Apoh
|
apolipoprotein H |
| chr1_+_165288606 | 0.55 |
ENSMUST00000027853.6
|
Mpc2
|
mitochondrial pyruvate carrier 2 |
| chr12_+_84332006 | 0.55 |
ENSMUST00000123614.8
ENSMUST00000147363.8 ENSMUST00000135001.8 ENSMUST00000146377.8 |
Ptgr2
|
prostaglandin reductase 2 |
| chr9_+_108167628 | 0.55 |
ENSMUST00000035227.8
|
Nicn1
|
nicolin 1 |
| chr12_+_8027767 | 0.54 |
ENSMUST00000037520.14
|
Apob
|
apolipoprotein B |
| chr14_-_6220396 | 0.54 |
ENSMUST00000224656.2
ENSMUST00000224154.2 ENSMUST00000022310.7 |
Ngly1
|
N-glycanase 1 |
| chr9_-_121745354 | 0.54 |
ENSMUST00000062474.5
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr5_+_122239030 | 0.53 |
ENSMUST00000139213.8
ENSMUST00000111751.8 ENSMUST00000155612.8 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr5_+_122239007 | 0.53 |
ENSMUST00000014080.13
ENSMUST00000111750.8 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr5_+_127709302 | 0.53 |
ENSMUST00000118139.3
|
Glt1d1
|
glycosyltransferase 1 domain containing 1 |
| chr7_-_119122681 | 0.52 |
ENSMUST00000033267.4
|
Pdilt
|
protein disulfide isomerase-like, testis expressed |
| chr19_-_39801188 | 0.52 |
ENSMUST00000162507.2
ENSMUST00000160476.9 ENSMUST00000239028.2 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chr5_+_45650821 | 0.52 |
ENSMUST00000198534.2
|
Lap3
|
leucine aminopeptidase 3 |
| chr13_+_24023386 | 0.51 |
ENSMUST00000039721.14
|
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chrX_-_48377072 | 0.50 |
ENSMUST00000114914.8
|
Enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr10_-_43880353 | 0.50 |
ENSMUST00000020017.14
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chrX_+_134786600 | 0.50 |
ENSMUST00000180025.8
ENSMUST00000148374.8 ENSMUST00000068755.14 |
Bhlhb9
|
basic helix-loop-helix domain containing, class B9 |
| chr10_-_125164399 | 0.50 |
ENSMUST00000063318.10
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
| chr1_-_150268470 | 0.49 |
ENSMUST00000006167.13
ENSMUST00000097547.10 |
Odr4
|
odr4 GPCR localization factor homolog |
| chr5_-_72716942 | 0.48 |
ENSMUST00000074948.5
ENSMUST00000087216.12 |
Nfxl1
|
nuclear transcription factor, X-box binding-like 1 |
| chr3_+_80943667 | 0.47 |
ENSMUST00000029652.4
|
Pdgfc
|
platelet-derived growth factor, C polypeptide |
| chr18_-_60860594 | 0.47 |
ENSMUST00000235795.2
|
Ndst1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr7_+_142052569 | 0.47 |
ENSMUST00000078497.15
ENSMUST00000105953.10 ENSMUST00000179658.8 ENSMUST00000105954.10 ENSMUST00000105952.10 ENSMUST00000105955.8 ENSMUST00000074187.13 ENSMUST00000169299.9 ENSMUST00000105957.10 ENSMUST00000180152.8 ENSMUST00000105950.11 ENSMUST00000105958.10 ENSMUST00000105949.8 |
Tnnt3
|
troponin T3, skeletal, fast |
| chr18_+_77032080 | 0.47 |
ENSMUST00000026485.15
ENSMUST00000150990.9 ENSMUST00000148955.3 |
Hdhd2
|
haloacid dehalogenase-like hydrolase domain containing 2 |
| chr10_-_125144678 | 0.46 |
ENSMUST00000105257.4
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
| chr2_+_15060051 | 0.46 |
ENSMUST00000069870.11
ENSMUST00000239125.2 ENSMUST00000193836.3 |
Arl5b
|
ADP-ribosylation factor-like 5B |
| chr4_+_129229373 | 0.46 |
ENSMUST00000141235.8
|
Zbtb8os
|
zinc finger and BTB domain containing 8 opposite strand |
| chr16_+_34842764 | 0.45 |
ENSMUST00000061156.10
|
Hacd2
|
3-hydroxyacyl-CoA dehydratase 2 |
| chr6_+_48653047 | 0.45 |
ENSMUST00000054050.5
|
Gimap9
|
GTPase, IMAP family member 9 |
| chr3_+_135531409 | 0.44 |
ENSMUST00000180196.8
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr11_+_21041291 | 0.44 |
ENSMUST00000093290.12
|
Peli1
|
pellino 1 |
| chr9_+_44966464 | 0.44 |
ENSMUST00000114664.8
|
Mpzl3
|
myelin protein zero-like 3 |
| chr4_-_107780716 | 0.43 |
ENSMUST00000106719.8
ENSMUST00000106720.9 ENSMUST00000131644.2 ENSMUST00000030345.15 |
Cpt2
|
carnitine palmitoyltransferase 2 |
| chr15_-_38976034 | 0.43 |
ENSMUST00000227323.2
ENSMUST00000022908.10 |
Slc25a32
|
solute carrier family 25, member 32 |
| chr6_+_121709891 | 0.43 |
ENSMUST00000204124.2
|
Gm7298
|
predicted gene 7298 |
| chr18_-_66425719 | 0.42 |
ENSMUST00000130300.3
|
Ccbe1
|
collagen and calcium binding EGF domains 1 |
| chr16_-_65359406 | 0.42 |
ENSMUST00000231259.2
|
Chmp2b
|
charged multivesicular body protein 2B |
| chr19_-_39875192 | 0.42 |
ENSMUST00000168838.3
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69 |
| chr15_-_78280099 | 0.41 |
ENSMUST00000229878.2
ENSMUST00000165170.8 ENSMUST00000074380.14 |
Tex33
|
testis expressed 33 |
| chr3_+_121085471 | 0.41 |
ENSMUST00000199554.2
|
Alg14
|
asparagine-linked glycosylation 14 |
| chr17_+_35643818 | 0.41 |
ENSMUST00000174699.8
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr5_+_93241287 | 0.41 |
ENSMUST00000074733.11
ENSMUST00000201700.4 ENSMUST00000202196.4 ENSMUST00000202308.4 |
Septin11
|
septin 11 |
| chrX_+_36059274 | 0.41 |
ENSMUST00000016463.4
|
Slc25a5
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 5 |
| chr3_+_121085373 | 0.39 |
ENSMUST00000039442.12
|
Alg14
|
asparagine-linked glycosylation 14 |
| chr18_+_56705894 | 0.39 |
ENSMUST00000008445.7
|
Phax
|
phosphorylated adaptor for RNA export |
| chr15_+_75881712 | 0.39 |
ENSMUST00000187868.3
|
Iqank1
|
IQ motif and ankyrin repeat containing 1 |
| chr7_-_48494959 | 0.39 |
ENSMUST00000208050.2
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr1_+_95241332 | 0.39 |
ENSMUST00000059975.8
ENSMUST00000186780.2 |
Fam174a
|
family with sequence similarity 174, member A |
| chr17_+_35643853 | 0.39 |
ENSMUST00000113879.4
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr14_+_26300693 | 0.39 |
ENSMUST00000203874.3
ENSMUST00000037585.9 |
Dennd6a
|
DENN/MADD domain containing 6A |
| chr17_+_75742881 | 0.38 |
ENSMUST00000164192.9
|
Rasgrp3
|
RAS, guanyl releasing protein 3 |
| chr7_-_103320398 | 0.38 |
ENSMUST00000062144.4
|
Olfr624
|
olfactory receptor 624 |
| chr2_-_101459274 | 0.38 |
ENSMUST00000099682.9
|
Iftap
|
intraflagellar transport associated protein |
| chr1_+_153300874 | 0.38 |
ENSMUST00000042373.12
|
Shcbp1l
|
Shc SH2-domain binding protein 1-like |
| chr5_+_147797258 | 0.37 |
ENSMUST00000031654.10
|
Pomp
|
proteasome maturation protein |
| chr14_+_53157900 | 0.37 |
ENSMUST00000178252.3
|
Trav9d-3
|
T cell receptor alpha variable 9D-3 |
| chr11_-_6425877 | 0.37 |
ENSMUST00000179343.3
|
Purb
|
purine rich element binding protein B |
| chr7_+_105053775 | 0.37 |
ENSMUST00000033187.6
ENSMUST00000210344.2 |
Cnga4
|
cyclic nucleotide gated channel alpha 4 |
| chr16_-_45664591 | 0.37 |
ENSMUST00000076333.12
|
Phldb2
|
pleckstrin homology like domain, family B, member 2 |
| chrX_-_36127891 | 0.37 |
ENSMUST00000115249.10
ENSMUST00000115248.10 |
C330007P06Rik
|
RIKEN cDNA C330007P06 gene |
| chr9_-_53617508 | 0.36 |
ENSMUST00000068449.4
|
Rab39
|
RAB39, member RAS oncogene family |
| chr3_-_73615535 | 0.36 |
ENSMUST00000138216.8
|
Bche
|
butyrylcholinesterase |
| chr5_+_32616187 | 0.36 |
ENSMUST00000015100.15
|
Ppp1cb
|
protein phosphatase 1 catalytic subunit beta |
| chr14_-_73613385 | 0.36 |
ENSMUST00000227454.2
|
Itm2b
|
integral membrane protein 2B |
| chr1_-_184578057 | 0.36 |
ENSMUST00000068725.10
|
Mtarc2
|
mitochondrial amidoxime reducing component 2 |
| chr6_-_48685108 | 0.36 |
ENSMUST00000126422.3
ENSMUST00000119315.2 ENSMUST00000053661.7 |
Gimap6
|
GTPase, IMAP family member 6 |
| chr9_+_73009680 | 0.36 |
ENSMUST00000034737.13
ENSMUST00000173734.9 ENSMUST00000167514.2 ENSMUST00000174203.3 |
Khdc3
Gm20509
|
KH domain containing 3, subcortical maternal complex member predicted gene 20509 |
| chr3_+_60380243 | 0.35 |
ENSMUST00000195724.6
|
Mbnl1
|
muscleblind like splicing factor 1 |
| chr1_-_106687457 | 0.35 |
ENSMUST00000010049.6
|
Kdsr
|
3-ketodihydrosphingosine reductase |
| chr7_+_65343156 | 0.35 |
ENSMUST00000032726.14
ENSMUST00000107495.5 ENSMUST00000143508.3 ENSMUST00000129166.3 ENSMUST00000206517.2 ENSMUST00000206837.2 ENSMUST00000206628.2 ENSMUST00000206361.2 |
Tm2d3
|
TM2 domain containing 3 |
| chr10_-_17898977 | 0.35 |
ENSMUST00000020002.9
|
Abracl
|
ABRA C-terminal like |
| chr12_-_84923252 | 0.35 |
ENSMUST00000163189.8
ENSMUST00000110254.9 ENSMUST00000002073.13 |
Ltbp2
|
latent transforming growth factor beta binding protein 2 |
| chr6_+_72332423 | 0.34 |
ENSMUST00000069695.9
ENSMUST00000132243.3 |
Tmem150a
|
transmembrane protein 150A |
| chr16_+_22710027 | 0.34 |
ENSMUST00000231848.2
|
Ahsg
|
alpha-2-HS-glycoprotein |
| chr4_-_96479793 | 0.34 |
ENSMUST00000055693.9
|
Cyp2j9
|
cytochrome P450, family 2, subfamily j, polypeptide 9 |
| chr14_-_68819544 | 0.34 |
ENSMUST00000022641.9
|
Adamdec1
|
ADAM-like, decysin 1 |
| chr13_-_14787602 | 0.34 |
ENSMUST00000220621.2
|
Mrpl32
|
mitochondrial ribosomal protein L32 |
| chr10_+_80165961 | 0.34 |
ENSMUST00000186864.7
ENSMUST00000040081.7 |
Reep6
|
receptor accessory protein 6 |
| chr5_-_38649291 | 0.34 |
ENSMUST00000129099.8
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr9_-_99450948 | 0.34 |
ENSMUST00000035043.12
|
Armc8
|
armadillo repeat containing 8 |
| chrX_-_74423647 | 0.33 |
ENSMUST00000114085.9
|
F8
|
coagulation factor VIII |
| chr4_-_49521036 | 0.33 |
ENSMUST00000057829.4
|
Mrpl50
|
mitochondrial ribosomal protein L50 |
| chr5_+_102629240 | 0.33 |
ENSMUST00000073302.12
ENSMUST00000094559.9 |
Arhgap24
|
Rho GTPase activating protein 24 |
| chr8_+_47246534 | 0.33 |
ENSMUST00000210218.2
|
Irf2
|
interferon regulatory factor 2 |
| chr6_-_6217021 | 0.33 |
ENSMUST00000015256.15
|
Slc25a13
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 13 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Dmc1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.6 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 1.1 | 3.2 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) |
| 1.0 | 3.1 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.6 | 1.7 | GO:0043387 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) |
| 0.6 | 1.7 | GO:1903173 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.5 | 6.2 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.5 | 1.5 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.5 | 1.9 | GO:0032379 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.5 | 1.0 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.5 | 1.4 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.4 | 5.6 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.4 | 5.5 | GO:0015747 | urate transport(GO:0015747) |
| 0.4 | 1.1 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.3 | 1.2 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.3 | 1.9 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.3 | 3.0 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.3 | 0.8 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.2 | 2.7 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.2 | 3.9 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.2 | 1.4 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.2 | 1.8 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 1.5 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.2 | 0.6 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.2 | 0.7 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.2 | 0.7 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.2 | 0.9 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.1 | 1.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 0.8 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.1 | 1.0 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.1 | 0.7 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 3.9 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.8 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.4 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.1 | 0.4 | GO:1903920 | positive regulation of actin filament severing(GO:1903920) |
| 0.1 | 0.6 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.7 | GO:0033668 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.1 | 0.9 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.1 | 1.7 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.3 | GO:2000536 | negative regulation of entry of bacterium into host cell(GO:2000536) |
| 0.1 | 0.8 | GO:0006545 | glycine biosynthetic process(GO:0006545) tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.1 | 0.3 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.1 | 0.1 | GO:0019585 | uronic acid metabolic process(GO:0006063) glucuronate metabolic process(GO:0019585) |
| 0.1 | 0.3 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.1 | 1.8 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.7 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.1 | 0.3 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.1 | 0.3 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.6 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 0.4 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 3.9 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.2 | GO:0090149 | synaptic vesicle recycling via endosome(GO:0036466) mitochondrial membrane fission(GO:0090149) regulation of peroxisome organization(GO:1900063) |
| 0.1 | 0.2 | GO:0042197 | dichloromethane metabolic process(GO:0018900) chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.1 | 1.0 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 2.5 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 1.3 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 0.4 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.5 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 0.4 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.1 | 0.1 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.1 | 0.1 | GO:0002590 | negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.1 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.1 | 0.3 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.1 | 0.4 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.2 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.6 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 0.4 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 0.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.2 | GO:1904156 | DN2 thymocyte differentiation(GO:1904155) DN3 thymocyte differentiation(GO:1904156) |
| 0.1 | 0.3 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 0.3 | GO:1904688 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.1 | 0.4 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 | 0.1 | GO:0006404 | RNA import into nucleus(GO:0006404) |
| 0.1 | 0.9 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 0.2 | GO:0030167 | proteoglycan catabolic process(GO:0030167) |
| 0.1 | 0.3 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 0.5 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.4 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 0.5 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 0.5 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.4 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.1 | 0.8 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 | 0.4 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.2 | GO:1904708 | paracrine signaling(GO:0038001) tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) granulosa cell apoptotic process(GO:1904700) regulation of granulosa cell apoptotic process(GO:1904708) |
| 0.0 | 0.3 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.0 | 0.8 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.9 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.5 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.2 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.8 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 1.3 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.2 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.1 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.0 | 0.5 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.4 | GO:0008063 | Toll signaling pathway(GO:0008063) positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.0 | 0.1 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 1.2 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.2 | GO:0072008 | glomerular mesangial cell differentiation(GO:0072008) glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.1 | GO:0002436 | immune complex clearance by monocytes and macrophages(GO:0002436) monocyte homeostasis(GO:0035702) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 2.8 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.1 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.0 | 0.6 | GO:0070255 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) regulation of mucus secretion(GO:0070255) |
| 0.0 | 0.5 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0009996 | negative regulation of cell fate specification(GO:0009996) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.6 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.0 | 0.1 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.3 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.4 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.2 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.1 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.0 | 0.1 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 | 0.1 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.0 | 5.3 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.1 | GO:0072347 | response to anesthetic(GO:0072347) |
| 0.0 | 0.4 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.2 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.1 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.0 | 0.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 1.9 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 0.2 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.1 | GO:0007522 | visceral muscle development(GO:0007522) |
| 0.0 | 0.4 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.3 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.1 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.0 | 0.1 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.4 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.8 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 2.7 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.9 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.5 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.0 | 0.2 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 0.4 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.1 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.1 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.1 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.0 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.0 | 0.2 | GO:2000232 | regulation of rRNA processing(GO:2000232) |
| 0.0 | 0.2 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) |
| 0.0 | 0.8 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.7 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.3 | GO:0001711 | endodermal cell fate commitment(GO:0001711) histone H2B ubiquitination(GO:0033523) |
| 0.0 | 1.0 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 1.2 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.1 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 0.2 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.3 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.0 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.0 | 0.2 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.1 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.1 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.1 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.0 | 0.4 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.5 | GO:0006505 | GPI anchor metabolic process(GO:0006505) GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.4 | GO:0071385 | cellular response to corticosteroid stimulus(GO:0071384) cellular response to glucocorticoid stimulus(GO:0071385) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 1.9 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.2 | 0.5 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.2 | 0.8 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.2 | 2.0 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 0.5 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 0.3 | GO:0044317 | rod spherule(GO:0044317) |
| 0.1 | 1.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.3 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 0.3 | GO:0034774 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.1 | 0.9 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 1.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.4 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 0.3 | GO:0034686 | integrin alphav-beta3 complex(GO:0034683) integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 0.6 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 1.9 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 1.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 1.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 4.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.4 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.5 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.8 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 10.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.7 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 2.7 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 1.3 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.1 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.0 | 0.6 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 0.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.2 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.1 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.7 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.2 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 1.4 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.2 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.7 | 2.6 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.7 | 2.0 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 0.5 | 1.5 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.5 | 1.9 | GO:1904121 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.5 | 1.4 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.4 | 1.7 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.4 | 1.1 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.4 | 14.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.3 | 5.8 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.3 | 2.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.3 | 1.3 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.3 | 1.5 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.3 | 0.9 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) single guanine insertion binding(GO:0032142) |
| 0.3 | 4.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.3 | 1.1 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.2 | 0.7 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.2 | 3.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.2 | 1.3 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.2 | 0.7 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.2 | 1.5 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.2 | 2.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 6.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.2 | 1.2 | GO:0030294 | receptor signaling protein tyrosine kinase inhibitor activity(GO:0030294) |
| 0.2 | 3.8 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.2 | 0.9 | GO:0030151 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) molybdenum ion binding(GO:0030151) |
| 0.2 | 0.7 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.2 | 3.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 0.5 | GO:0080023 | 3R-hydroxyacyl-CoA dehydratase activity(GO:0080023) |
| 0.1 | 0.7 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.1 | 0.6 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.1 | 0.7 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.1 | 0.8 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.6 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.5 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 0.8 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.1 | 0.8 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 1.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.4 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.1 | 0.3 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 0.6 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.6 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 0.2 | GO:0019120 | hydrolase activity, acting on acid halide bonds(GO:0016824) hydrolase activity, acting on acid halide bonds, in C-halide compounds(GO:0019120) alkylhalidase activity(GO:0047651) |
| 0.1 | 0.5 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.1 | 1.8 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 0.4 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.9 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.1 | 0.8 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.1 | 0.2 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 0.4 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 0.6 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.9 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.2 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.1 | 1.0 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.1 | 0.2 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 0.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 1.2 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 0.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.3 | GO:0052901 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.1 | 0.2 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 0.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 1.0 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.7 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.5 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 1.9 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 1.4 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.0 | 0.3 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.8 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.5 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.3 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.0 | 0.8 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.2 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.0 | 1.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.4 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.2 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.0 | 0.1 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.1 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 1.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 1.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.4 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.0 | 1.4 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.0 | 0.6 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.7 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.2 | GO:0052724 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 1.0 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.7 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.2 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.3 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.5 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 1.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.5 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0047874 | dolichyldiphosphatase activity(GO:0047874) |
| 0.0 | 0.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 1.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 2.3 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.1 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 2.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.5 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.0 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.1 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.6 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.9 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 4.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 1.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 2.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 5.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 3.9 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.3 | 0.8 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.2 | 0.7 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.2 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 2.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 1.9 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 2.8 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 3.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 0.8 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 0.4 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.1 | 1.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.8 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 1.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.0 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 3.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.7 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.5 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.4 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.9 | REACTOME PI3K EVENTS IN ERBB2 SIGNALING | Genes involved in PI3K events in ERBB2 signaling |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |