Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
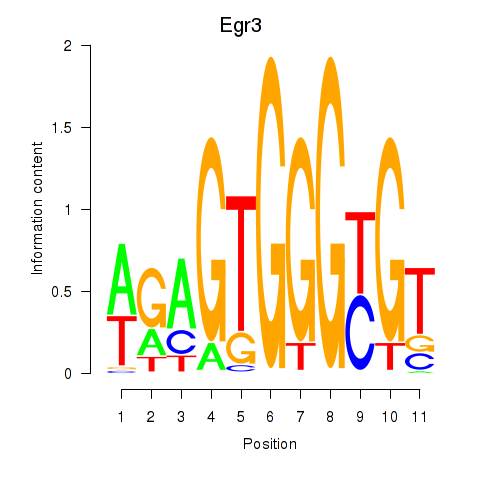
Results for Egr3
Z-value: 0.72
Transcription factors associated with Egr3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Egr3
|
ENSMUSG00000033730.5 | early growth response 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Egr3 | mm39_v1_chr14_+_70314652_70314671 | -0.16 | 3.5e-01 | Click! |
Activity profile of Egr3 motif
Sorted Z-values of Egr3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_94063823 | 5.19 |
ENSMUST00000044602.8
|
Ces1g
|
carboxylesterase 1G |
| chr14_+_66205932 | 4.03 |
ENSMUST00000022616.14
|
Clu
|
clusterin |
| chr12_-_84497718 | 3.79 |
ENSMUST00000085192.7
ENSMUST00000220491.2 |
Aldh6a1
|
aldehyde dehydrogenase family 6, subfamily A1 |
| chr1_+_133291302 | 3.26 |
ENSMUST00000135222.9
|
Etnk2
|
ethanolamine kinase 2 |
| chr6_-_124519240 | 3.07 |
ENSMUST00000159463.8
ENSMUST00000162844.2 ENSMUST00000160505.8 ENSMUST00000162443.8 |
C1s1
|
complement component 1, s subcomponent 1 |
| chr10_-_68114543 | 2.77 |
ENSMUST00000219238.2
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like) |
| chr17_+_25798059 | 2.58 |
ENSMUST00000141606.3
ENSMUST00000063344.15 ENSMUST00000116641.9 |
Lmf1
|
lipase maturation factor 1 |
| chr11_-_98666159 | 2.44 |
ENSMUST00000064941.7
|
Nr1d1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr5_-_134776101 | 2.33 |
ENSMUST00000015138.13
|
Eln
|
elastin |
| chr4_-_129132963 | 1.95 |
ENSMUST00000097873.10
|
C77080
|
expressed sequence C77080 |
| chr19_-_38113249 | 1.65 |
ENSMUST00000112335.4
|
Rbp4
|
retinol binding protein 4, plasma |
| chr11_-_69586626 | 1.56 |
ENSMUST00000108649.3
ENSMUST00000174159.8 ENSMUST00000181810.8 |
Tnfsfm13
Tnfsf12
|
tumor necrosis factor (ligand) superfamily, membrane-bound member 13 tumor necrosis factor (ligand) superfamily, member 12 |
| chr7_-_30754792 | 1.54 |
ENSMUST00000206328.2
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr10_+_106306122 | 1.51 |
ENSMUST00000029404.17
ENSMUST00000217854.2 |
Ppfia2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr19_-_38113056 | 1.47 |
ENSMUST00000236283.2
|
Rbp4
|
retinol binding protein 4, plasma |
| chr7_+_34818709 | 1.40 |
ENSMUST00000205391.2
ENSMUST00000042985.11 |
Cebpa
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr11_+_4833186 | 1.38 |
ENSMUST00000139737.2
|
Nipsnap1
|
nipsnap homolog 1 |
| chr6_+_91661074 | 1.10 |
ENSMUST00000205480.2
ENSMUST00000206545.2 |
Slc6a6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
| chr11_-_68277631 | 1.09 |
ENSMUST00000021284.4
|
Ntn1
|
netrin 1 |
| chr8_+_119010458 | 1.05 |
ENSMUST00000117160.2
|
Cdh13
|
cadherin 13 |
| chr10_-_59057570 | 0.96 |
ENSMUST00000220156.2
ENSMUST00000165971.3 |
Septin10
|
septin 10 |
| chrX_-_47602395 | 0.93 |
ENSMUST00000114945.9
ENSMUST00000037349.8 |
Aifm1
|
apoptosis-inducing factor, mitochondrion-associated 1 |
| chr2_-_121244251 | 0.91 |
ENSMUST00000038073.5
|
Catsper2
|
cation channel, sperm associated 2 |
| chr2_+_121244256 | 0.88 |
ENSMUST00000028683.14
|
Pdia3
|
protein disulfide isomerase associated 3 |
| chr13_+_59733163 | 0.87 |
ENSMUST00000166923.9
|
Naa35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
| chr11_-_109613040 | 0.86 |
ENSMUST00000020938.8
|
Fam20a
|
FAM20A, golgi associated secretory pathway pseudokinase |
| chr18_-_38068456 | 0.85 |
ENSMUST00000080033.7
ENSMUST00000115631.8 ENSMUST00000115634.8 |
Diaph1
|
diaphanous related formin 1 |
| chr10_-_127504416 | 0.82 |
ENSMUST00000129252.2
|
Nab2
|
Ngfi-A binding protein 2 |
| chr18_-_38068430 | 0.82 |
ENSMUST00000025337.14
|
Diaph1
|
diaphanous related formin 1 |
| chr16_-_94171853 | 0.80 |
ENSMUST00000113914.8
ENSMUST00000113905.8 |
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr13_-_85437201 | 0.79 |
ENSMUST00000223598.2
|
Rasa1
|
RAS p21 protein activator 1 |
| chr5_+_65127412 | 0.73 |
ENSMUST00000031080.15
|
Fam114a1
|
family with sequence similarity 114, member A1 |
| chr2_-_58457168 | 0.73 |
ENSMUST00000056376.12
|
Acvr1
|
activin A receptor, type 1 |
| chr4_+_57637817 | 0.71 |
ENSMUST00000150412.4
|
Pakap
|
paralemmin A kinase anchor protein |
| chr13_-_85437118 | 0.69 |
ENSMUST00000109552.3
|
Rasa1
|
RAS p21 protein activator 1 |
| chr2_-_143853122 | 0.69 |
ENSMUST00000016072.12
ENSMUST00000037875.6 |
Rrbp1
|
ribosome binding protein 1 |
| chr16_-_45664664 | 0.67 |
ENSMUST00000036355.13
|
Phldb2
|
pleckstrin homology like domain, family B, member 2 |
| chr16_-_45664591 | 0.67 |
ENSMUST00000076333.12
|
Phldb2
|
pleckstrin homology like domain, family B, member 2 |
| chr7_+_43361930 | 0.63 |
ENSMUST00000066834.8
|
Klk13
|
kallikrein related-peptidase 13 |
| chr18_-_84104507 | 0.61 |
ENSMUST00000060303.10
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr5_+_138278502 | 0.60 |
ENSMUST00000160729.8
|
Stag3
|
stromal antigen 3 |
| chr18_-_84104574 | 0.59 |
ENSMUST00000175783.3
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr4_+_40948407 | 0.57 |
ENSMUST00000030128.6
|
Chmp5
|
charged multivesicular body protein 5 |
| chr9_-_50639367 | 0.56 |
ENSMUST00000117646.8
|
Dixdc1
|
DIX domain containing 1 |
| chr7_+_48608800 | 0.56 |
ENSMUST00000183659.8
|
Nav2
|
neuron navigator 2 |
| chr13_+_59733073 | 0.55 |
ENSMUST00000168367.8
ENSMUST00000022038.15 |
Naa35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
| chr10_+_80134917 | 0.52 |
ENSMUST00000154212.8
|
Apc2
|
APC regulator of WNT signaling pathway 2 |
| chr10_-_17898838 | 0.51 |
ENSMUST00000220433.2
|
Abracl
|
ABRA C-terminal like |
| chr7_-_45159739 | 0.51 |
ENSMUST00000211682.2
ENSMUST00000033096.16 ENSMUST00000209436.2 ENSMUST00000211343.2 |
Nucb1
|
nucleobindin 1 |
| chrX_+_142447361 | 0.51 |
ENSMUST00000126592.8
ENSMUST00000156449.8 ENSMUST00000155215.8 ENSMUST00000112865.8 |
Pak3
|
p21 (RAC1) activated kinase 3 |
| chrX_+_35459621 | 0.49 |
ENSMUST00000115256.2
|
Zcchc12
|
zinc finger, CCHC domain containing 12 |
| chr17_-_46638874 | 0.45 |
ENSMUST00000047970.14
|
Abcc10
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 10 |
| chrX_+_142447286 | 0.44 |
ENSMUST00000112868.8
|
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr5_-_21850579 | 0.44 |
ENSMUST00000051358.11
|
Fbxl13
|
F-box and leucine-rich repeat protein 13 |
| chr11_-_68277799 | 0.44 |
ENSMUST00000135141.2
|
Ntn1
|
netrin 1 |
| chr10_+_40759815 | 0.43 |
ENSMUST00000105509.2
|
Wasf1
|
WASP family, member 1 |
| chr5_-_138278223 | 0.42 |
ENSMUST00000014089.9
ENSMUST00000161827.8 |
Gpc2
|
glypican 2 (cerebroglycan) |
| chr5_-_62923463 | 0.40 |
ENSMUST00000076623.8
ENSMUST00000159470.3 |
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr1_+_158190090 | 0.39 |
ENSMUST00000194369.6
ENSMUST00000195311.6 |
Astn1
|
astrotactin 1 |
| chr16_-_94171556 | 0.39 |
ENSMUST00000113906.9
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr10_+_40759468 | 0.37 |
ENSMUST00000019975.14
|
Wasf1
|
WASP family, member 1 |
| chr7_+_144450260 | 0.36 |
ENSMUST00000033389.7
ENSMUST00000207229.2 |
Fgf15
|
fibroblast growth factor 15 |
| chr2_+_27055245 | 0.36 |
ENSMUST00000000910.7
|
Dbh
|
dopamine beta hydroxylase |
| chr17_-_46638915 | 0.35 |
ENSMUST00000167360.8
|
Abcc10
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 10 |
| chr3_+_93301003 | 0.34 |
ENSMUST00000045912.3
|
Rptn
|
repetin |
| chr11_-_99313078 | 0.33 |
ENSMUST00000017741.4
|
Krt12
|
keratin 12 |
| chr10_+_3784877 | 0.32 |
ENSMUST00000239159.2
|
Plekhg1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr15_-_98676007 | 0.32 |
ENSMUST00000226655.2
ENSMUST00000228546.2 ENSMUST00000023732.12 ENSMUST00000226610.2 |
Wnt10b
|
wingless-type MMTV integration site family, member 10B |
| chr10_+_83558729 | 0.32 |
ENSMUST00000150459.3
|
1500009L16Rik
|
RIKEN cDNA 1500009L16 gene |
| chr3_+_106943472 | 0.32 |
ENSMUST00000052718.5
|
Kcna3
|
potassium voltage-gated channel, shaker-related subfamily, member 3 |
| chr16_+_21023505 | 0.31 |
ENSMUST00000006112.7
|
Ephb3
|
Eph receptor B3 |
| chr12_+_49429790 | 0.31 |
ENSMUST00000021333.5
|
Foxg1
|
forkhead box G1 |
| chr16_-_94171533 | 0.30 |
ENSMUST00000113910.8
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr9_-_54956032 | 0.29 |
ENSMUST00000034854.8
|
Chrnb4
|
cholinergic receptor, nicotinic, beta polypeptide 4 |
| chr11_-_89587671 | 0.29 |
ENSMUST00000128717.9
|
Ankfn1
|
ankyrin-repeat and fibronectin type III domain containing 1 |
| chr19_+_6276408 | 0.29 |
ENSMUST00000025698.14
ENSMUST00000113526.2 |
Gpha2
|
glycoprotein hormone alpha 2 |
| chr19_+_59447950 | 0.29 |
ENSMUST00000174353.2
|
Emx2
|
empty spiracles homeobox 2 |
| chr7_-_45159790 | 0.29 |
ENSMUST00000211765.2
|
Nucb1
|
nucleobindin 1 |
| chr10_-_17898938 | 0.27 |
ENSMUST00000220110.2
|
Abracl
|
ABRA C-terminal like |
| chr2_-_69416365 | 0.27 |
ENSMUST00000100051.9
ENSMUST00000092551.5 ENSMUST00000080953.12 |
Lrp2
|
low density lipoprotein receptor-related protein 2 |
| chr1_+_120268299 | 0.26 |
ENSMUST00000037286.12
|
C1ql2
|
complement component 1, q subcomponent-like 2 |
| chr4_-_20778530 | 0.25 |
ENSMUST00000119374.8
|
Nkain3
|
Na+/K+ transporting ATPase interacting 3 |
| chr9_-_58111589 | 0.25 |
ENSMUST00000217578.2
ENSMUST00000114144.9 ENSMUST00000214649.2 |
Islr
Islr2
|
immunoglobulin superfamily containing leucine-rich repeat immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr14_-_21790463 | 0.24 |
ENSMUST00000120984.9
|
Dusp13
|
dual specificity phosphatase 13 |
| chrX_+_159945990 | 0.24 |
ENSMUST00000077375.5
|
Scml2
|
Scm polycomb group protein like 2 |
| chr1_-_164135008 | 0.24 |
ENSMUST00000027866.11
ENSMUST00000120447.8 ENSMUST00000086032.4 |
Blzf1
|
basic leucine zipper nuclear factor 1 |
| chr17_-_80022480 | 0.23 |
ENSMUST00000234361.2
|
Cyp1b1
|
cytochrome P450, family 1, subfamily b, polypeptide 1 |
| chr17_-_80022463 | 0.23 |
ENSMUST00000024894.2
|
Cyp1b1
|
cytochrome P450, family 1, subfamily b, polypeptide 1 |
| chr11_+_71641806 | 0.22 |
ENSMUST00000108511.8
|
Wscd1
|
WSC domain containing 1 |
| chr11_+_71641505 | 0.21 |
ENSMUST00000021168.14
|
Wscd1
|
WSC domain containing 1 |
| chrX_+_159945740 | 0.21 |
ENSMUST00000074802.12
ENSMUST00000019101.11 ENSMUST00000112345.8 |
Scml2
|
Scm polycomb group protein like 2 |
| chr8_-_49008881 | 0.20 |
ENSMUST00000110345.8
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr6_-_83549399 | 0.20 |
ENSMUST00000206592.2
ENSMUST00000206400.2 |
Stambp
|
STAM binding protein |
| chr9_+_21077010 | 0.19 |
ENSMUST00000039413.15
|
Pde4a
|
phosphodiesterase 4A, cAMP specific |
| chr7_+_18810167 | 0.19 |
ENSMUST00000108479.2
|
Dmwd
|
dystrophia myotonica-containing WD repeat motif |
| chr7_+_3648264 | 0.19 |
ENSMUST00000206287.2
ENSMUST00000038913.16 |
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr14_+_55747902 | 0.19 |
ENSMUST00000165262.8
ENSMUST00000074225.11 |
Cpne6
|
copine VI |
| chr15_-_78428865 | 0.18 |
ENSMUST00000053239.4
|
Sstr3
|
somatostatin receptor 3 |
| chr7_+_80510658 | 0.17 |
ENSMUST00000132163.8
ENSMUST00000205361.2 ENSMUST00000147125.2 |
Zscan2
|
zinc finger and SCAN domain containing 2 |
| chr19_-_38212544 | 0.17 |
ENSMUST00000067167.6
|
Fra10ac1
|
FRA10AC1 homolog (human) |
| chr12_+_49429574 | 0.16 |
ENSMUST00000179669.3
|
Foxg1
|
forkhead box G1 |
| chr10_-_40759307 | 0.15 |
ENSMUST00000044166.9
|
Cdc40
|
cell division cycle 40 |
| chr11_-_107805830 | 0.15 |
ENSMUST00000039071.3
|
Cacng5
|
calcium channel, voltage-dependent, gamma subunit 5 |
| chr11_-_103158190 | 0.14 |
ENSMUST00000021324.3
|
Map3k14
|
mitogen-activated protein kinase kinase kinase 14 |
| chr10_-_17898977 | 0.14 |
ENSMUST00000020002.9
|
Abracl
|
ABRA C-terminal like |
| chr7_-_118783820 | 0.13 |
ENSMUST00000084650.6
|
Gpr139
|
G protein-coupled receptor 139 |
| chr17_+_87590308 | 0.12 |
ENSMUST00000041110.12
ENSMUST00000125875.8 |
Ttc7
|
tetratricopeptide repeat domain 7 |
| chr1_-_161615927 | 0.11 |
ENSMUST00000193648.2
|
Fasl
|
Fas ligand (TNF superfamily, member 6) |
| chr1_-_9369463 | 0.11 |
ENSMUST00000140295.8
|
Sntg1
|
syntrophin, gamma 1 |
| chr16_-_88087087 | 0.11 |
ENSMUST00000211444.2
ENSMUST00000023652.16 ENSMUST00000072256.13 |
Grik1
|
glutamate receptor, ionotropic, kainate 1 |
| chr8_+_63404395 | 0.08 |
ENSMUST00000119068.8
|
Spock3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 3 |
| chr3_-_126955976 | 0.08 |
ENSMUST00000182994.8
|
Ank2
|
ankyrin 2, brain |
| chr9_+_119723931 | 0.08 |
ENSMUST00000036561.8
ENSMUST00000217472.2 ENSMUST00000215307.2 |
Wdr48
|
WD repeat domain 48 |
| chr15_+_99122742 | 0.06 |
ENSMUST00000041415.5
|
Kcnh3
|
potassium voltage-gated channel, subfamily H (eag-related), member 3 |
| chr3_+_105359641 | 0.06 |
ENSMUST00000098761.10
|
Kcnd3
|
potassium voltage-gated channel, Shal-related family, member 3 |
| chr1_-_161616031 | 0.06 |
ENSMUST00000000834.4
|
Fasl
|
Fas ligand (TNF superfamily, member 6) |
| chr4_+_32238950 | 0.05 |
ENSMUST00000037416.13
|
Bach2
|
BTB and CNC homology, basic leucine zipper transcription factor 2 |
| chr9_+_55056648 | 0.04 |
ENSMUST00000121677.8
|
Ube2q2
|
ubiquitin-conjugating enzyme E2Q family member 2 |
| chr5_+_138278777 | 0.04 |
ENSMUST00000048028.15
ENSMUST00000162245.8 ENSMUST00000161691.2 |
Stag3
|
stromal antigen 3 |
| chr4_+_99812912 | 0.04 |
ENSMUST00000102783.5
|
Pgm1
|
phosphoglucomutase 1 |
| chr12_+_86999366 | 0.03 |
ENSMUST00000191463.2
|
Cipc
|
CLOCK interacting protein, circadian |
| chr8_+_63404228 | 0.03 |
ENSMUST00000118003.8
|
Spock3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 3 |
| chr6_-_88851579 | 0.02 |
ENSMUST00000061262.11
ENSMUST00000140455.8 ENSMUST00000145780.2 |
Podxl2
|
podocalyxin-like 2 |
| chr16_+_94171477 | 0.01 |
ENSMUST00000117648.9
ENSMUST00000147352.8 ENSMUST00000150346.8 ENSMUST00000155692.8 ENSMUST00000153988.9 ENSMUST00000139513.9 ENSMUST00000141856.8 ENSMUST00000152117.8 ENSMUST00000150097.8 ENSMUST00000122895.8 ENSMUST00000151770.8 ENSMUST00000231569.2 ENSMUST00000147046.8 ENSMUST00000149885.8 ENSMUST00000127667.8 ENSMUST00000119131.3 ENSMUST00000145883.2 |
Ttc3
|
tetratricopeptide repeat domain 3 |
| chr9_-_79869299 | 0.00 |
ENSMUST00000172973.2
|
Filip1
|
filamin A interacting protein 1 |
| chr3_+_97808506 | 0.00 |
ENSMUST00000029476.9
ENSMUST00000130620.2 ENSMUST00000122288.2 |
Sec22b
|
SEC22 homolog B, vesicle trafficking protein |
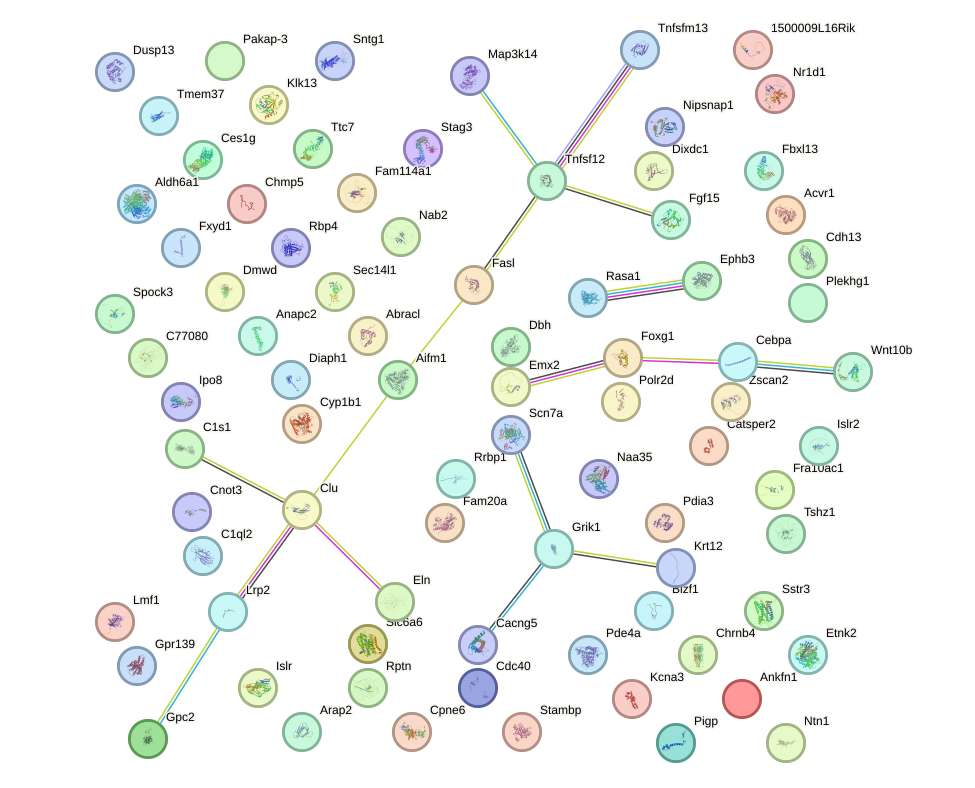
Network of associatons between targets according to the STRING database.
First level regulatory network of Egr3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0090320 | chylomicron assembly(GO:0034378) regulation of chylomicron remnant clearance(GO:0090320) |
| 0.9 | 3.8 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.7 | 4.0 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.6 | 3.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.5 | 2.4 | GO:0070859 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.4 | 2.6 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.4 | 1.1 | GO:0001762 | beta-alanine transport(GO:0001762) taurine transport(GO:0015734) |
| 0.3 | 3.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.3 | 0.9 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.2 | 3.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 2.8 | GO:0060613 | fat pad development(GO:0060613) |
| 0.2 | 0.6 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.2 | 1.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.2 | 0.8 | GO:1902949 | positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.2 | 0.5 | GO:0021852 | pyramidal neuron migration(GO:0021852) |
| 0.2 | 0.5 | GO:0002930 | trabecular meshwork development(GO:0002930) endothelial cell-cell adhesion(GO:0071603) |
| 0.2 | 1.1 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.1 | 0.7 | GO:0003274 | endocardial cushion fusion(GO:0003274) positive regulation of determination of dorsal identity(GO:2000017) |
| 0.1 | 1.5 | GO:1903278 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.1 | 1.5 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 1.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.6 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.1 | 1.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.4 | GO:0006589 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.1 | 1.4 | GO:0000050 | urea cycle(GO:0000050) |
| 0.1 | 0.6 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.8 | GO:1990416 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 0.3 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.1 | 1.5 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.1 | 0.9 | GO:1902510 | regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.1 | 0.3 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.1 | 0.4 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.4 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.1 | 0.9 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 1.4 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.6 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.2 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 | 1.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.3 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.9 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 1.0 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.0 | 0.5 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.3 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.0 | 1.6 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.1 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.0 | 0.3 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.3 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.3 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.5 | 2.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.4 | 1.4 | GO:0031417 | NatC complex(GO:0031417) |
| 0.3 | 4.0 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.2 | 0.7 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.8 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.1 | 0.6 | GO:0000802 | transverse filament(GO:0000802) |
| 0.1 | 0.4 | GO:0034774 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.1 | 0.9 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 1.5 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 1.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.9 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 1.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 1.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 1.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.4 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 3.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.3 | GO:0001533 | cornified envelope(GO:0001533) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.8 | 3.3 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.4 | 1.1 | GO:0005368 | beta-alanine transmembrane transporter activity(GO:0001761) taurine transmembrane transporter activity(GO:0005368) taurine:sodium symporter activity(GO:0005369) |
| 0.3 | 0.9 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.2 | 0.9 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.2 | 1.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.2 | 4.0 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.4 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.1 | 1.5 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.7 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 2.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.3 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.1 | 0.3 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 1.4 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.1 | 1.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 3.8 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.1 | 1.5 | GO:0043422 | platelet-derived growth factor receptor binding(GO:0005161) protein kinase B binding(GO:0043422) |
| 0.0 | 2.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 1.7 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.0 | 1.0 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 1.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 5.2 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.9 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.0 | 0.9 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 1.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.4 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.6 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 3.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.5 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.5 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.5 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 4.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 0.7 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 5.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.5 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.9 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 2.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 3.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.5 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 2.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 0.9 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 2.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |