Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
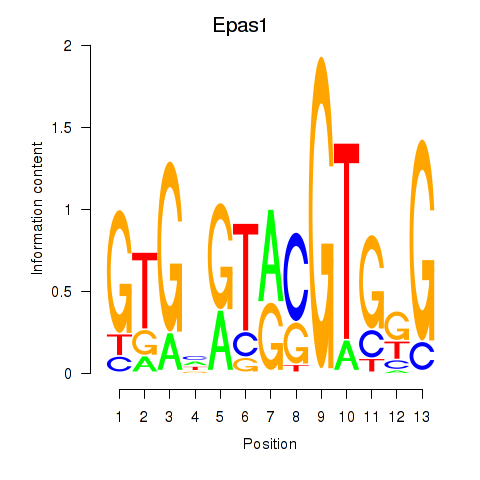
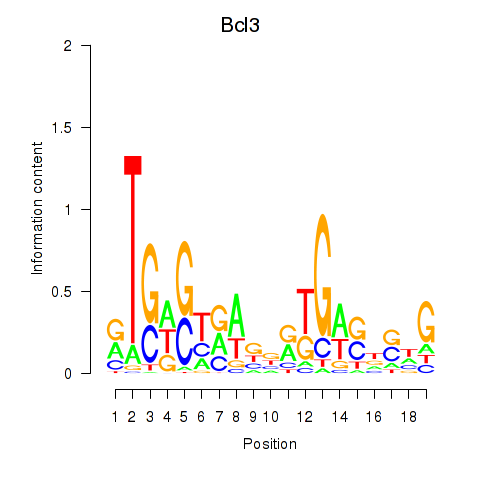
Results for Epas1_Bcl3
Z-value: 1.13
Transcription factors associated with Epas1_Bcl3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Epas1
|
ENSMUSG00000024140.11 | endothelial PAS domain protein 1 |
|
Bcl3
|
ENSMUSG00000053175.18 | B cell leukemia/lymphoma 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bcl3 | mm39_v1_chr7_-_19556612_19556712 | 0.74 | 3.1e-07 | Click! |
| Epas1 | mm39_v1_chr17_+_87061117_87061137 | 0.45 | 5.9e-03 | Click! |
Activity profile of Epas1_Bcl3 motif
Sorted Z-values of Epas1_Bcl3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_25872836 | 9.56 |
ENSMUST00000082214.5
|
Cyp2b9
|
cytochrome P450, family 2, subfamily b, polypeptide 9 |
| chr15_-_3612078 | 7.57 |
ENSMUST00000161770.2
|
Ghr
|
growth hormone receptor |
| chr7_-_30754792 | 7.02 |
ENSMUST00000206328.2
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr8_+_114860297 | 5.97 |
ENSMUST00000073521.12
ENSMUST00000066514.13 |
Nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr5_-_134776101 | 4.79 |
ENSMUST00000015138.13
|
Eln
|
elastin |
| chr17_-_35100980 | 4.71 |
ENSMUST00000152417.8
ENSMUST00000146299.8 |
C2
Gm20547
|
complement component 2 (within H-2S) predicted gene 20547 |
| chr1_-_90897329 | 4.47 |
ENSMUST00000130042.2
ENSMUST00000027529.12 |
Rab17
|
RAB17, member RAS oncogene family |
| chr1_-_192923816 | 4.21 |
ENSMUST00000160929.8
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr17_-_84990360 | 3.60 |
ENSMUST00000066175.10
|
Abcg5
|
ATP binding cassette subfamily G member 5 |
| chr17_-_35101069 | 3.54 |
ENSMUST00000025230.15
|
C2
|
complement component 2 (within H-2S) |
| chr10_+_87694117 | 3.51 |
ENSMUST00000122386.8
|
Igf1
|
insulin-like growth factor 1 |
| chr19_+_8641369 | 3.46 |
ENSMUST00000035444.10
ENSMUST00000163785.2 |
Chrm1
|
cholinergic receptor, muscarinic 1, CNS |
| chr7_-_98010478 | 3.30 |
ENSMUST00000094161.11
ENSMUST00000164726.8 ENSMUST00000206414.2 ENSMUST00000167405.3 |
Tsku
|
tsukushi, small leucine rich proteoglycan |
| chr7_+_25760922 | 3.26 |
ENSMUST00000005669.9
|
Cyp2b13
|
cytochrome P450, family 2, subfamily b, polypeptide 13 |
| chr16_-_23339548 | 3.17 |
ENSMUST00000089883.7
|
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr11_+_98239230 | 3.17 |
ENSMUST00000078694.13
|
Ppp1r1b
|
protein phosphatase 1, regulatory inhibitor subunit 1B |
| chr17_+_84990541 | 3.07 |
ENSMUST00000045714.15
ENSMUST00000171915.2 |
Abcg8
|
ATP binding cassette subfamily G member 8 |
| chr11_+_112673041 | 3.00 |
ENSMUST00000000579.3
|
Sox9
|
SRY (sex determining region Y)-box 9 |
| chr8_-_112356957 | 2.85 |
ENSMUST00000070004.4
|
Ldhd
|
lactate dehydrogenase D |
| chr6_+_71176811 | 2.71 |
ENSMUST00000067492.8
|
Fabp1
|
fatty acid binding protein 1, liver |
| chr17_-_28779678 | 2.70 |
ENSMUST00000114785.3
ENSMUST00000025062.5 |
Clps
|
colipase, pancreatic |
| chr6_+_91661034 | 2.66 |
ENSMUST00000032185.9
|
Slc6a6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
| chr17_-_35077089 | 2.65 |
ENSMUST00000153400.8
|
Cfb
|
complement factor B |
| chr7_-_98010534 | 2.64 |
ENSMUST00000165257.8
|
Tsku
|
tsukushi, small leucine rich proteoglycan |
| chr6_+_40605758 | 2.45 |
ENSMUST00000202636.4
ENSMUST00000201148.4 ENSMUST00000071535.10 |
Mgam
|
maltase-glucoamylase |
| chr8_-_85526653 | 2.39 |
ENSMUST00000126806.2
ENSMUST00000076715.13 |
Nfix
|
nuclear factor I/X |
| chr7_-_140590605 | 2.36 |
ENSMUST00000026565.7
|
Ifitm3
|
interferon induced transmembrane protein 3 |
| chr4_-_46991842 | 2.30 |
ENSMUST00000107749.4
|
Gabbr2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr11_+_98932586 | 2.28 |
ENSMUST00000177092.8
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr6_-_84570890 | 2.27 |
ENSMUST00000168003.9
|
Cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr6_+_91661074 | 2.25 |
ENSMUST00000205480.2
ENSMUST00000206545.2 |
Slc6a6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
| chr9_+_108174052 | 2.22 |
ENSMUST00000035230.7
|
Amt
|
aminomethyltransferase |
| chr2_-_165997179 | 2.15 |
ENSMUST00000088086.4
|
Sulf2
|
sulfatase 2 |
| chr7_+_100970435 | 2.13 |
ENSMUST00000210192.2
ENSMUST00000172630.8 |
Stard10
|
START domain containing 10 |
| chr1_+_166081664 | 2.12 |
ENSMUST00000111416.7
|
Ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr6_+_121323577 | 2.06 |
ENSMUST00000032200.16
|
Slc6a12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 |
| chr15_-_83054369 | 2.05 |
ENSMUST00000162834.3
|
Cyb5r3
|
cytochrome b5 reductase 3 |
| chr12_-_103979900 | 1.98 |
ENSMUST00000058464.5
|
Serpina9
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 9 |
| chr2_+_169474916 | 1.98 |
ENSMUST00000109159.3
|
Tshz2
|
teashirt zinc finger family member 2 |
| chr5_-_77262968 | 1.97 |
ENSMUST00000081964.7
|
Hopx
|
HOP homeobox |
| chr2_-_52566583 | 1.97 |
ENSMUST00000178799.8
|
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr5_-_92653377 | 1.93 |
ENSMUST00000031377.9
|
Scarb2
|
scavenger receptor class B, member 2 |
| chr6_+_88701578 | 1.93 |
ENSMUST00000150180.4
ENSMUST00000163271.8 |
Mgll
|
monoglyceride lipase |
| chr18_-_38999755 | 1.86 |
ENSMUST00000115582.8
ENSMUST00000236060.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr7_-_30755007 | 1.85 |
ENSMUST00000206474.2
ENSMUST00000205807.2 ENSMUST00000039909.13 ENSMUST00000206305.2 ENSMUST00000205439.2 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr4_+_137589548 | 1.84 |
ENSMUST00000102518.10
|
Ece1
|
endothelin converting enzyme 1 |
| chr16_-_23339329 | 1.82 |
ENSMUST00000230040.2
ENSMUST00000229619.2 |
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr16_-_38533597 | 1.82 |
ENSMUST00000023487.5
|
Arhgap31
|
Rho GTPase activating protein 31 |
| chr16_-_97723753 | 1.80 |
ENSMUST00000170757.3
|
C2cd2
|
C2 calcium-dependent domain containing 2 |
| chrX_-_160942713 | 1.79 |
ENSMUST00000087085.10
|
Nhs
|
NHS actin remodeling regulator |
| chr7_+_100970910 | 1.78 |
ENSMUST00000174291.8
ENSMUST00000167888.9 ENSMUST00000172662.2 |
Stard10
|
START domain containing 10 |
| chr19_+_23736205 | 1.78 |
ENSMUST00000025830.9
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chr7_+_100971034 | 1.74 |
ENSMUST00000173270.8
|
Stard10
|
START domain containing 10 |
| chr3_-_30563611 | 1.74 |
ENSMUST00000173899.8
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr9_-_50639230 | 1.73 |
ENSMUST00000118707.2
ENSMUST00000034566.15 |
Dixdc1
|
DIX domain containing 1 |
| chr6_+_124470053 | 1.73 |
ENSMUST00000049124.10
|
C1rl
|
complement component 1, r subcomponent-like |
| chr15_-_96953823 | 1.68 |
ENSMUST00000023101.10
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr3_+_94280101 | 1.65 |
ENSMUST00000029795.10
|
Rorc
|
RAR-related orphan receptor gamma |
| chr1_+_166081755 | 1.65 |
ENSMUST00000194964.6
ENSMUST00000192638.6 ENSMUST00000192426.6 ENSMUST00000195557.6 ENSMUST00000192732.6 ENSMUST00000193860.2 |
Ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr5_-_8672951 | 1.64 |
ENSMUST00000047485.15
ENSMUST00000115378.2 |
Rundc3b
|
RUN domain containing 3B |
| chr3_-_137837117 | 1.64 |
ENSMUST00000029805.13
|
Mttp
|
microsomal triglyceride transfer protein |
| chr5_-_73495895 | 1.63 |
ENSMUST00000202012.2
|
Ociad2
|
OCIA domain containing 2 |
| chr4_-_129132963 | 1.63 |
ENSMUST00000097873.10
|
C77080
|
expressed sequence C77080 |
| chr9_+_121471782 | 1.61 |
ENSMUST00000035115.5
|
Vipr1
|
vasoactive intestinal peptide receptor 1 |
| chr8_-_85526972 | 1.55 |
ENSMUST00000099070.10
|
Nfix
|
nuclear factor I/X |
| chr10_-_108846816 | 1.55 |
ENSMUST00000105276.8
ENSMUST00000064054.14 |
Syt1
|
synaptotagmin I |
| chr16_+_23338960 | 1.51 |
ENSMUST00000211460.2
ENSMUST00000210658.2 ENSMUST00000209198.2 ENSMUST00000210371.2 ENSMUST00000211499.2 ENSMUST00000210795.2 ENSMUST00000209422.2 |
Gm45338
Rtp4
|
predicted gene 45338 receptor transporter protein 4 |
| chr8_+_119010458 | 1.49 |
ENSMUST00000117160.2
|
Cdh13
|
cadherin 13 |
| chrX_-_51254129 | 1.44 |
ENSMUST00000033450.3
|
Gpc4
|
glypican 4 |
| chr7_-_25331168 | 1.40 |
ENSMUST00000205808.2
|
Bckdha
|
branched chain ketoacid dehydrogenase E1, alpha polypeptide |
| chr1_+_152275575 | 1.33 |
ENSMUST00000044311.9
|
Colgalt2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr12_-_103925197 | 1.32 |
ENSMUST00000122229.8
|
Serpina1e
|
serine (or cysteine) peptidase inhibitor, clade A, member 1E |
| chr11_-_116545071 | 1.29 |
ENSMUST00000021166.6
|
Cygb
|
cytoglobin |
| chr8_+_77626400 | 1.28 |
ENSMUST00000109913.9
|
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr10_-_71180763 | 1.26 |
ENSMUST00000045887.9
|
Cisd1
|
CDGSH iron sulfur domain 1 |
| chr7_+_86895851 | 1.26 |
ENSMUST00000032781.14
|
Nox4
|
NADPH oxidase 4 |
| chr7_+_119499322 | 1.25 |
ENSMUST00000106516.2
|
Lyrm1
|
LYR motif containing 1 |
| chr7_-_100306160 | 1.23 |
ENSMUST00000107046.8
ENSMUST00000107045.9 ENSMUST00000139708.9 |
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr14_+_118374511 | 1.22 |
ENSMUST00000022728.4
|
Gpr180
|
G protein-coupled receptor 180 |
| chr11_-_101676076 | 1.19 |
ENSMUST00000164750.8
ENSMUST00000107176.8 ENSMUST00000017868.7 |
Etv4
|
ets variant 4 |
| chr2_-_165996716 | 1.15 |
ENSMUST00000139266.2
|
Sulf2
|
sulfatase 2 |
| chr5_+_102629365 | 1.10 |
ENSMUST00000112854.8
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr1_-_191129223 | 1.09 |
ENSMUST00000067976.9
|
Ppp2r5a
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr18_-_77652820 | 1.07 |
ENSMUST00000026494.14
ENSMUST00000182024.2 |
Rnf165
|
ring finger protein 165 |
| chr9_+_44394080 | 1.05 |
ENSMUST00000220303.2
|
Bcl9l
|
B cell CLL/lymphoma 9-like |
| chr8_-_3744167 | 1.04 |
ENSMUST00000005678.6
|
Fcer2a
|
Fc receptor, IgE, low affinity II, alpha polypeptide |
| chr15_+_81686622 | 1.02 |
ENSMUST00000109553.10
|
Tef
|
thyrotroph embryonic factor |
| chr8_-_49008881 | 1.02 |
ENSMUST00000110345.8
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr9_-_50639367 | 1.02 |
ENSMUST00000117646.8
|
Dixdc1
|
DIX domain containing 1 |
| chr12_-_103409912 | 1.01 |
ENSMUST00000055071.9
|
Ifi27l2a
|
interferon, alpha-inducible protein 27 like 2A |
| chr4_-_42661893 | 0.96 |
ENSMUST00000108006.4
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr17_-_13087012 | 0.96 |
ENSMUST00000089015.10
|
Mas1
|
MAS1 oncogene |
| chr11_-_69586347 | 0.96 |
ENSMUST00000181261.2
|
Tnfsf12
|
tumor necrosis factor (ligand) superfamily, member 12 |
| chr1_+_42734051 | 0.96 |
ENSMUST00000239323.2
ENSMUST00000199521.5 ENSMUST00000176807.3 |
Pou3f3
Gm20646
|
POU domain, class 3, transcription factor 3 predicted gene 20646 |
| chr4_+_85972125 | 0.95 |
ENSMUST00000107178.9
ENSMUST00000048885.12 ENSMUST00000141889.8 ENSMUST00000120678.2 |
Adamtsl1
|
ADAMTS-like 1 |
| chr4_-_131871797 | 0.95 |
ENSMUST00000056336.2
|
Oprd1
|
opioid receptor, delta 1 |
| chr11_-_113600346 | 0.95 |
ENSMUST00000173655.8
ENSMUST00000100248.6 |
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr11_+_108811168 | 0.95 |
ENSMUST00000052915.14
|
Axin2
|
axin 2 |
| chr5_+_24633206 | 0.95 |
ENSMUST00000115049.9
|
Slc4a2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr11_+_60308077 | 0.94 |
ENSMUST00000070681.7
|
Gid4
|
GID complex subunit 4, VID24 homolog |
| chr13_+_42205790 | 0.94 |
ENSMUST00000220525.2
|
Hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr7_+_101027390 | 0.93 |
ENSMUST00000084895.12
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr7_-_44465043 | 0.92 |
ENSMUST00000107893.9
|
Atf5
|
activating transcription factor 5 |
| chr10_-_127587576 | 0.92 |
ENSMUST00000079692.6
|
Gpr182
|
G protein-coupled receptor 182 |
| chr8_+_47246534 | 0.92 |
ENSMUST00000210218.2
|
Irf2
|
interferon regulatory factor 2 |
| chr7_-_141119703 | 0.90 |
ENSMUST00000118694.8
ENSMUST00000153191.8 ENSMUST00000026586.13 ENSMUST00000166082.8 |
Chid1
|
chitinase domain containing 1 |
| chr19_+_26725589 | 0.90 |
ENSMUST00000207812.2
ENSMUST00000175791.9 ENSMUST00000207118.2 ENSMUST00000209085.2 ENSMUST00000112637.10 ENSMUST00000207054.2 ENSMUST00000208589.2 ENSMUST00000176475.9 ENSMUST00000176698.9 ENSMUST00000207832.2 ENSMUST00000177252.9 ENSMUST00000208712.2 ENSMUST00000208186.2 ENSMUST00000208806.2 ENSMUST00000208027.2 |
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr7_-_140676596 | 0.88 |
ENSMUST00000209199.2
|
Sigirr
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr18_+_64473091 | 0.88 |
ENSMUST00000175965.10
|
Onecut2
|
one cut domain, family member 2 |
| chr2_-_69416365 | 0.88 |
ENSMUST00000100051.9
ENSMUST00000092551.5 ENSMUST00000080953.12 |
Lrp2
|
low density lipoprotein receptor-related protein 2 |
| chrX_+_150127171 | 0.88 |
ENSMUST00000073364.6
|
Fam120c
|
family with sequence similarity 120, member C |
| chr13_+_58955675 | 0.87 |
ENSMUST00000224402.2
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr11_+_28803188 | 0.87 |
ENSMUST00000020759.12
|
Efemp1
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 1 |
| chr18_-_33597060 | 0.86 |
ENSMUST00000168890.2
|
Nrep
|
neuronal regeneration related protein |
| chr6_+_15185562 | 0.85 |
ENSMUST00000137628.8
|
Foxp2
|
forkhead box P2 |
| chr7_-_140535899 | 0.85 |
ENSMUST00000081649.10
|
Ifitm2
|
interferon induced transmembrane protein 2 |
| chr9_-_106769131 | 0.83 |
ENSMUST00000159283.8
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr9_-_106769069 | 0.83 |
ENSMUST00000160503.4
ENSMUST00000159620.9 |
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr2_+_119181703 | 0.83 |
ENSMUST00000028780.4
|
Chac1
|
ChaC, cation transport regulator 1 |
| chr17_-_24866749 | 0.82 |
ENSMUST00000234121.2
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr11_+_49135018 | 0.82 |
ENSMUST00000167400.8
ENSMUST00000081794.7 |
Mgat1
|
mannoside acetylglucosaminyltransferase 1 |
| chr5_-_144902598 | 0.82 |
ENSMUST00000110677.8
ENSMUST00000085684.11 ENSMUST00000100461.7 |
Smurf1
|
SMAD specific E3 ubiquitin protein ligase 1 |
| chr3_-_107839133 | 0.82 |
ENSMUST00000004137.11
|
Gstm7
|
glutathione S-transferase, mu 7 |
| chr14_+_122116002 | 0.79 |
ENSMUST00000039803.7
|
Ubac2
|
ubiquitin associated domain containing 2 |
| chr11_+_97690391 | 0.79 |
ENSMUST00000043843.12
|
Lasp1
|
LIM and SH3 protein 1 |
| chr10_+_106306122 | 0.79 |
ENSMUST00000029404.17
ENSMUST00000217854.2 |
Ppfia2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr14_-_100522101 | 0.79 |
ENSMUST00000228216.2
|
Klf12
|
Kruppel-like factor 12 |
| chr12_+_4284009 | 0.79 |
ENSMUST00000179139.3
|
Ptrhd1
|
peptidyl-tRNA hydrolase domain containing 1 |
| chr8_+_3637785 | 0.78 |
ENSMUST00000171962.3
ENSMUST00000207712.2 ENSMUST00000207970.2 ENSMUST00000207533.2 ENSMUST00000208240.2 ENSMUST00000207432.2 ENSMUST00000207077.2 |
Camsap3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr10_+_39488930 | 0.77 |
ENSMUST00000019987.7
|
Traf3ip2
|
TRAF3 interacting protein 2 |
| chr6_+_88701394 | 0.77 |
ENSMUST00000113585.9
|
Mgll
|
monoglyceride lipase |
| chr6_+_15185399 | 0.74 |
ENSMUST00000115474.8
ENSMUST00000115472.8 ENSMUST00000031545.14 |
Foxp2
|
forkhead box P2 |
| chr17_-_87573294 | 0.73 |
ENSMUST00000145895.8
ENSMUST00000129616.8 ENSMUST00000155904.2 ENSMUST00000151155.8 ENSMUST00000144236.9 ENSMUST00000024963.11 |
Mcfd2
|
multiple coagulation factor deficiency 2 |
| chr18_-_25886908 | 0.73 |
ENSMUST00000115816.3
ENSMUST00000223704.2 |
Celf4
|
CUGBP, Elav-like family member 4 |
| chr17_+_49239393 | 0.72 |
ENSMUST00000046254.3
|
Lrfn2
|
leucine rich repeat and fibronectin type III domain containing 2 |
| chrX_-_94209913 | 0.72 |
ENSMUST00000113873.9
ENSMUST00000113876.9 ENSMUST00000199920.5 ENSMUST00000113885.8 ENSMUST00000113883.8 ENSMUST00000196012.2 ENSMUST00000182001.8 ENSMUST00000113878.8 ENSMUST00000113882.8 ENSMUST00000182562.2 |
Arhgef9
|
CDC42 guanine nucleotide exchange factor (GEF) 9 |
| chr15_+_59520493 | 0.71 |
ENSMUST00000118228.2
|
Trib1
|
tribbles pseudokinase 1 |
| chr2_+_68691902 | 0.71 |
ENSMUST00000176018.2
|
Cers6
|
ceramide synthase 6 |
| chr9_+_21279802 | 0.70 |
ENSMUST00000214474.2
|
Ilf3
|
interleukin enhancer binding factor 3 |
| chr12_-_14202041 | 0.70 |
ENSMUST00000020926.8
|
Lratd1
|
LRAT domain containing 1 |
| chr13_+_31990604 | 0.69 |
ENSMUST00000062292.5
|
Foxc1
|
forkhead box C1 |
| chr11_-_118460736 | 0.69 |
ENSMUST00000136551.3
|
Rbfox3
|
RNA binding protein, fox-1 homolog (C. elegans) 3 |
| chr11_-_113600838 | 0.67 |
ENSMUST00000018871.8
|
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr4_-_97666279 | 0.67 |
ENSMUST00000146447.8
|
E130114P18Rik
|
RIKEN cDNA E130114P18 gene |
| chr15_+_82159398 | 0.66 |
ENSMUST00000023095.14
ENSMUST00000230365.2 |
Septin3
|
septin 3 |
| chr9_+_62765362 | 0.66 |
ENSMUST00000213643.2
ENSMUST00000034777.14 ENSMUST00000163820.3 ENSMUST00000215870.2 ENSMUST00000214633.2 ENSMUST00000215968.2 |
Calml4
|
calmodulin-like 4 |
| chr3_+_105359641 | 0.66 |
ENSMUST00000098761.10
|
Kcnd3
|
potassium voltage-gated channel, Shal-related family, member 3 |
| chr4_+_48045143 | 0.65 |
ENSMUST00000030025.10
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr1_+_180878797 | 0.65 |
ENSMUST00000036819.7
|
9130409I23Rik
|
RIKEN cDNA 9130409I23 gene |
| chr10_+_127478844 | 0.65 |
ENSMUST00000092074.12
ENSMUST00000120279.2 |
Stat6
|
signal transducer and activator of transcription 6 |
| chr12_-_101784727 | 0.64 |
ENSMUST00000222587.2
|
Fbln5
|
fibulin 5 |
| chr4_-_141351110 | 0.64 |
ENSMUST00000038661.8
|
Slc25a34
|
solute carrier family 25, member 34 |
| chr9_+_20779924 | 0.64 |
ENSMUST00000043911.8
|
Shfl
|
shiftless antiviral inhibitor of ribosomal frameshifting |
| chr11_-_118103540 | 0.64 |
ENSMUST00000106302.9
ENSMUST00000151165.2 |
Cyth1
|
cytohesin 1 |
| chr3_-_30563919 | 0.63 |
ENSMUST00000172697.8
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr2_-_63014514 | 0.63 |
ENSMUST00000112452.2
|
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr11_+_50917831 | 0.61 |
ENSMUST00000072152.2
|
Olfr54
|
olfactory receptor 54 |
| chr6_-_124888643 | 0.61 |
ENSMUST00000032217.2
|
Lag3
|
lymphocyte-activation gene 3 |
| chr9_-_21996693 | 0.61 |
ENSMUST00000179422.8
ENSMUST00000098937.10 ENSMUST00000177967.2 ENSMUST00000180180.8 |
Ecsit
|
ECSIT signalling integrator |
| chr5_-_136003294 | 0.60 |
ENSMUST00000154181.2
ENSMUST00000111152.8 ENSMUST00000111153.8 |
Ssc4d
|
scavenger receptor cysteine rich family, 4 domains |
| chr4_+_97665992 | 0.59 |
ENSMUST00000107062.9
ENSMUST00000052018.12 ENSMUST00000107057.8 |
Nfia
|
nuclear factor I/A |
| chr9_+_26645024 | 0.59 |
ENSMUST00000160899.8
ENSMUST00000161431.3 ENSMUST00000159799.8 |
B3gat1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) |
| chr15_+_34837501 | 0.59 |
ENSMUST00000072868.5
|
Kcns2
|
K+ voltage-gated channel, subfamily S, 2 |
| chr1_-_14374842 | 0.58 |
ENSMUST00000188857.7
ENSMUST00000185453.7 |
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr13_+_19427015 | 0.58 |
ENSMUST00000198663.5
ENSMUST00000103559.3 |
Trgv3
|
T cell receptor gamma, variable 3 |
| chr13_+_58955506 | 0.58 |
ENSMUST00000079828.7
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr5_-_116584765 | 0.58 |
ENSMUST00000139425.2
|
Srrm4
|
serine/arginine repetitive matrix 4 |
| chrX_-_47602395 | 0.58 |
ENSMUST00000114945.9
ENSMUST00000037349.8 |
Aifm1
|
apoptosis-inducing factor, mitochondrion-associated 1 |
| chr11_+_95010935 | 0.57 |
ENSMUST00000092768.7
|
Dlx3
|
distal-less homeobox 3 |
| chr18_-_25886750 | 0.56 |
ENSMUST00000224553.2
ENSMUST00000025117.14 |
Celf4
|
CUGBP, Elav-like family member 4 |
| chr8_+_109429732 | 0.56 |
ENSMUST00000188994.2
|
Zfhx3
|
zinc finger homeobox 3 |
| chr6_+_127049865 | 0.56 |
ENSMUST00000000186.9
|
Fgf23
|
fibroblast growth factor 23 |
| chr14_-_100521888 | 0.56 |
ENSMUST00000226774.2
|
Klf12
|
Kruppel-like factor 12 |
| chr17_+_44263890 | 0.56 |
ENSMUST00000177857.9
ENSMUST00000044792.6 |
Rcan2
|
regulator of calcineurin 2 |
| chr6_-_29179583 | 0.56 |
ENSMUST00000159200.2
|
Prrt4
|
proline-rich transmembrane protein 4 |
| chr11_-_60307847 | 0.56 |
ENSMUST00000108721.9
ENSMUST00000239016.2 |
Atpaf2
|
ATP synthase mitochondrial F1 complex assembly factor 2 |
| chr1_+_66361252 | 0.56 |
ENSMUST00000123647.8
|
Map2
|
microtubule-associated protein 2 |
| chr15_-_37458768 | 0.55 |
ENSMUST00000116445.9
|
Ncald
|
neurocalcin delta |
| chr18_-_25887173 | 0.55 |
ENSMUST00000225477.2
|
Celf4
|
CUGBP, Elav-like family member 4 |
| chr8_-_123980908 | 0.54 |
ENSMUST00000122363.8
|
Vps9d1
|
VPS9 domain containing 1 |
| chr6_+_15185202 | 0.53 |
ENSMUST00000154448.2
|
Foxp2
|
forkhead box P2 |
| chr2_+_173579285 | 0.53 |
ENSMUST00000067530.6
|
Vapb
|
vesicle-associated membrane protein, associated protein B and C |
| chr3_-_131196213 | 0.53 |
ENSMUST00000197057.2
|
Sgms2
|
sphingomyelin synthase 2 |
| chr10_-_127504416 | 0.53 |
ENSMUST00000129252.2
|
Nab2
|
Ngfi-A binding protein 2 |
| chr5_+_35915217 | 0.52 |
ENSMUST00000101280.10
ENSMUST00000054598.12 ENSMUST00000114205.8 ENSMUST00000114206.9 |
Ablim2
|
actin-binding LIM protein 2 |
| chr7_+_130294403 | 0.52 |
ENSMUST00000207282.2
|
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr2_+_29717354 | 0.52 |
ENSMUST00000091142.4
|
Urm1
|
ubiquitin related modifier 1 |
| chr9_-_102231884 | 0.51 |
ENSMUST00000035129.14
ENSMUST00000085169.12 ENSMUST00000149800.3 |
Ephb1
|
Eph receptor B1 |
| chr1_-_174749379 | 0.51 |
ENSMUST00000055294.4
|
Grem2
|
gremlin 2, DAN family BMP antagonist |
| chr7_+_95859332 | 0.50 |
ENSMUST00000138760.8
|
Tenm4
|
teneurin transmembrane protein 4 |
| chr2_+_163662752 | 0.50 |
ENSMUST00000029188.8
|
Ccn5
|
cellular communication network factor 5 |
| chr13_+_89687915 | 0.49 |
ENSMUST00000022108.9
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr5_+_65127412 | 0.49 |
ENSMUST00000031080.15
|
Fam114a1
|
family with sequence similarity 114, member A1 |
| chr13_+_119565118 | 0.49 |
ENSMUST00000109203.9
|
Paip1
|
polyadenylate binding protein-interacting protein 1 |
| chr2_-_118378020 | 0.49 |
ENSMUST00000110859.3
|
Bmf
|
BCL2 modifying factor |
| chr6_-_101176147 | 0.49 |
ENSMUST00000239140.2
|
Pdzrn3
|
PDZ domain containing RING finger 3 |
| chr3_-_148696155 | 0.48 |
ENSMUST00000196526.5
ENSMUST00000200543.5 ENSMUST00000200154.5 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr3_-_127292636 | 0.48 |
ENSMUST00000189368.2
|
Ank2
|
ankyrin 2, brain |
| chr5_+_130477642 | 0.47 |
ENSMUST00000111288.4
|
Caln1
|
calneuron 1 |
| chr9_+_26645141 | 0.47 |
ENSMUST00000115269.9
|
B3gat1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Epas1_Bcl3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:0001762 | beta-alanine transport(GO:0001762) taurine transport(GO:0015734) |
| 1.5 | 3.0 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 1.5 | 6.0 | GO:0033869 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) acetyl-CoA catabolic process(GO:0046356) |
| 1.5 | 4.5 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 1.4 | 4.2 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 1.2 | 5.9 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 1.1 | 6.7 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.9 | 8.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.8 | 8.9 | GO:1903797 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.7 | 2.2 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.7 | 7.6 | GO:0000255 | allantoin metabolic process(GO:0000255) isoleucine metabolic process(GO:0006549) |
| 0.7 | 3.4 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.6 | 3.2 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.6 | 1.8 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.6 | 5.6 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.6 | 1.7 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.5 | 3.3 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.5 | 3.5 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.4 | 2.7 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.4 | 2.1 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.4 | 1.6 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.4 | 12.8 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.4 | 5.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.4 | 2.3 | GO:2001037 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.3 | 1.3 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.3 | 0.9 | GO:1904092 | regulation of autophagic cell death(GO:1904092) negative regulation of autophagic cell death(GO:1904093) |
| 0.3 | 0.8 | GO:0071211 | protein targeting to vacuole involved in autophagy(GO:0071211) |
| 0.3 | 1.9 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.2 | 1.5 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.2 | 1.0 | GO:0072023 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.2 | 0.7 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.2 | 0.6 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.2 | 1.5 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.2 | 2.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.2 | 0.8 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.2 | 3.6 | GO:0048537 | mucosal-associated lymphoid tissue development(GO:0048537) Peyer's patch development(GO:0048541) |
| 0.2 | 3.9 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 2.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.2 | 2.9 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.2 | 2.0 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.2 | 0.9 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.2 | 2.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 0.7 | GO:1901491 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) negative regulation of lymphangiogenesis(GO:1901491) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.2 | 1.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 2.7 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.2 | 0.5 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.2 | 0.7 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.2 | 2.1 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.2 | 0.5 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.2 | 0.9 | GO:2000054 | regulation of chondrocyte development(GO:0061181) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 1.0 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.1 | 1.6 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 1.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.4 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.1 | 0.6 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.1 | 0.8 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.1 | 1.0 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 0.5 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) negative regulation by virus of viral protein levels in host cell(GO:0046725) |
| 0.1 | 0.5 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.1 | 0.5 | GO:0014719 | skeletal muscle satellite cell activation(GO:0014719) |
| 0.1 | 0.5 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.1 | 0.4 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 1.2 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 | 0.8 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.1 | 0.5 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.1 | 0.6 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.9 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.6 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 0.6 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 | 0.8 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 1.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 2.8 | GO:0060013 | righting reflex(GO:0060013) |
| 0.1 | 1.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.5 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 | 2.0 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.9 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.1 | 0.9 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 0.6 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.1 | 2.8 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 0.3 | GO:0034443 | regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.1 | 0.4 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.1 | 0.5 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 1.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 0.8 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 0.5 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.1 | 0.4 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 1.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.2 | GO:0014862 | regulation of the force of skeletal muscle contraction(GO:0014728) regulation of skeletal muscle contraction by chemo-mechanical energy conversion(GO:0014862) |
| 0.1 | 0.4 | GO:1904457 | glucosylceramide catabolic process(GO:0006680) positive regulation of protein lipidation(GO:1903061) positive regulation of neuronal action potential(GO:1904457) |
| 0.1 | 1.8 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 | 1.4 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 0.4 | GO:0002266 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.1 | 0.4 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 0.6 | GO:1902510 | regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.1 | 0.4 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.2 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.1 | 2.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 3.8 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 | 1.7 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.1 | 0.3 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.1 | 0.6 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.2 | GO:0033577 | protein glycosylation in endoplasmic reticulum(GO:0033577) |
| 0.1 | 0.9 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 1.1 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 0.2 | GO:0003274 | B cell tolerance induction(GO:0002514) regulation of tolerance induction to self antigen(GO:0002649) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) endocardial cushion fusion(GO:0003274) positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.1 | 0.3 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.1 | 0.6 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.1 | 1.0 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 0.4 | GO:0044334 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.1 | 0.8 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 0.2 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.1 | 0.6 | GO:2000121 | regulation of removal of superoxide radicals(GO:2000121) |
| 0.1 | 0.3 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) |
| 0.1 | 2.6 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 0.2 | GO:1904155 | DN2 thymocyte differentiation(GO:1904155) DN3 thymocyte differentiation(GO:1904156) |
| 0.1 | 0.2 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.1 | 0.3 | GO:0090335 | regulation of brown fat cell differentiation(GO:0090335) |
| 0.1 | 0.4 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 0.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.2 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.6 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 1.5 | GO:0030204 | chondroitin sulfate metabolic process(GO:0030204) |
| 0.1 | 0.4 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.7 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.0 | 1.3 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 1.1 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 1.0 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 1.1 | GO:0060384 | innervation(GO:0060384) |
| 0.0 | 1.2 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.8 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.1 | GO:0021934 | hindbrain tangential cell migration(GO:0021934) |
| 0.0 | 2.9 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.0 | 0.2 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.1 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.0 | 6.1 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.9 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.4 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.0 | 0.8 | GO:0001783 | B cell apoptotic process(GO:0001783) |
| 0.0 | 0.2 | GO:0071313 | cellular response to caffeine(GO:0071313) sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.0 | 0.2 | GO:0035696 | monocyte extravasation(GO:0035696) |
| 0.0 | 0.1 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.0 | 2.1 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 0.3 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.2 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.3 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 1.0 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.2 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 1.4 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.1 | GO:0061081 | positive regulation of myeloid leukocyte cytokine production involved in immune response(GO:0061081) |
| 0.0 | 0.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.4 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.3 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.3 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.3 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.5 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.2 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.0 | 0.2 | GO:0034391 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 | 0.0 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.0 | 0.5 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:0021658 | rhombomere 3 morphogenesis(GO:0021658) |
| 0.0 | 0.1 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.1 | GO:0098907 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.0 | 0.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) osteoclast fusion(GO:0072675) |
| 0.0 | 0.7 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.7 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.2 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.7 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.6 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.1 | GO:0061143 | alveolar primary septum development(GO:0061143) paramesonephric duct development(GO:0061205) |
| 0.0 | 0.6 | GO:0051341 | regulation of oxidoreductase activity(GO:0051341) |
| 0.0 | 0.2 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 1.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.2 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 1.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:0015819 | lysine transport(GO:0015819) |
| 0.0 | 0.4 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.4 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 2.8 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 0.3 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.4 | GO:0010107 | potassium ion import(GO:0010107) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 1.1 | 5.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.5 | 2.3 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.5 | 2.3 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.5 | 6.9 | GO:0098533 | ATPase dependent transmembrane transport complex(GO:0098533) |
| 0.4 | 3.5 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.3 | 5.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 2.7 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 1.4 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.2 | 1.8 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.2 | 3.4 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 9.1 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 1.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 4.5 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 2.7 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 1.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.4 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 1.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.4 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 0.4 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 0.6 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.4 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 0.8 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.4 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 1.6 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 1.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 1.0 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 3.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.5 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 1.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 2.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 6.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.7 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.2 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 1.0 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.6 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 4.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 2.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 5.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.9 | GO:0031526 | brush border membrane(GO:0031526) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 1.6 | 4.9 | GO:0005368 | beta-alanine transmembrane transporter activity(GO:0001761) taurine transmembrane transporter activity(GO:0005368) taurine:sodium symporter activity(GO:0005369) |
| 1.4 | 4.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 1.2 | 6.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 1.1 | 3.2 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 1.0 | 2.9 | GO:0016898 | D-lactate dehydrogenase (cytochrome) activity(GO:0004458) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.7 | 3.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.5 | 1.6 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.5 | 3.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.5 | 1.4 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.4 | 1.3 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.4 | 2.4 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.4 | 1.6 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.4 | 2.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.4 | 2.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.3 | 12.8 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.3 | 1.3 | GO:0004096 | catalase activity(GO:0004096) |
| 0.3 | 2.7 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.3 | 0.6 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.3 | 2.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.3 | 0.9 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.3 | 1.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 1.5 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.2 | 1.5 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.2 | 3.0 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.2 | 2.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 0.9 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.2 | 1.5 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.2 | 6.7 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.2 | 2.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.2 | 0.8 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.2 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.2 | 0.5 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.2 | 1.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.2 | 2.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.2 | 2.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 0.7 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.2 | 1.0 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.2 | 0.8 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.2 | 0.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.4 | GO:0099530 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 0.8 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.4 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.1 | 0.7 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 1.0 | GO:0019863 | IgE binding(GO:0019863) |
| 0.1 | 0.4 | GO:0052692 | raffinose alpha-galactosidase activity(GO:0052692) |
| 0.1 | 0.4 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 1.0 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.1 | 9.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.6 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 5.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.9 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.1 | 0.9 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.5 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 0.5 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 3.9 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.3 | GO:0042008 | interleukin-18 receptor activity(GO:0042008) |
| 0.1 | 1.9 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 1.8 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 1.6 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 2.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.4 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.7 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 0.2 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.1 | 0.6 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.2 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 12.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 0.4 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 3.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.9 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) ankyrin repeat binding(GO:0071532) |
| 0.0 | 1.3 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.1 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 0.0 | 0.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.9 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.2 | GO:0019962 | interferon receptor activity(GO:0004904) type I interferon receptor activity(GO:0004905) type I interferon binding(GO:0019962) |
| 0.0 | 0.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 1.3 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.8 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 1.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.5 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 1.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.7 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 1.8 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.5 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.8 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 2.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.6 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.3 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.6 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 1.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 3.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 1.0 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.0 | GO:0071820 | N-box binding(GO:0071820) |
| 0.0 | 1.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 1.2 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 3.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 12.3 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) |
| 0.0 | 0.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.2 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 2.0 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.3 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 7.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 2.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 3.9 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 0.9 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 10.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 1.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.0 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.9 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.0 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.7 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 7.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.7 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 2.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.1 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.5 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.5 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 15.7 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.6 | 2.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.3 | 6.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.3 | 6.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.3 | 7.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.2 | 7.8 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 1.6 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 2.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 2.4 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 2.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 2.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 2.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 3.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 3.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.6 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 4.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 3.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.7 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 1.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 2.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.0 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 3.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.9 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 1.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.8 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 2.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 1.1 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 2.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.1 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |