Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Esr1
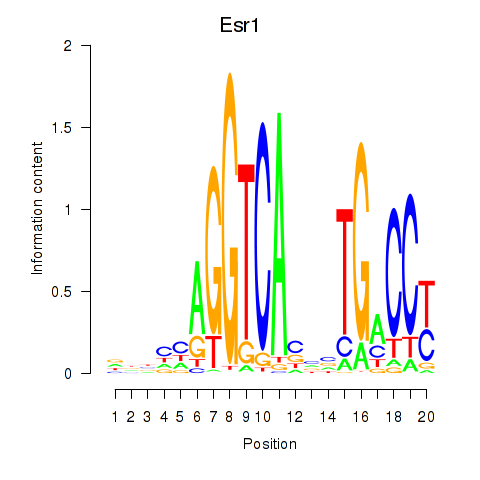
Z-value: 1.90
Transcription factors associated with Esr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Esr1
|
ENSMUSG00000019768.17 | estrogen receptor 1 (alpha) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Esr1 | mm39_v1_chr10_+_4561974_4562075 | -0.72 | 9.5e-07 | Click! |
Activity profile of Esr1 motif
Sorted Z-values of Esr1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_110848339 | 21.16 |
ENSMUST00000198884.5
ENSMUST00000196777.5 ENSMUST00000196209.5 ENSMUST00000035077.8 ENSMUST00000196122.3 |
Ltf
|
lactotransferrin |
| chr11_-_102255999 | 20.23 |
ENSMUST00000006749.10
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr10_+_79722081 | 19.13 |
ENSMUST00000046091.7
|
Elane
|
elastase, neutrophil expressed |
| chr11_+_87685032 | 17.34 |
ENSMUST00000121303.8
|
Mpo
|
myeloperoxidase |
| chr11_+_87684299 | 16.41 |
ENSMUST00000020779.11
|
Mpo
|
myeloperoxidase |
| chr1_+_131566044 | 14.72 |
ENSMUST00000073350.13
|
Ctse
|
cathepsin E |
| chr1_+_131566223 | 13.87 |
ENSMUST00000112411.2
|
Ctse
|
cathepsin E |
| chr11_+_58808830 | 12.57 |
ENSMUST00000020792.12
ENSMUST00000108818.4 |
Btnl10
|
butyrophilin-like 10 |
| chr11_-_83177548 | 11.12 |
ENSMUST00000163961.3
|
Slfn14
|
schlafen 14 |
| chr9_-_70328816 | 10.94 |
ENSMUST00000034742.8
|
Ccnb2
|
cyclin B2 |
| chr4_-_119047202 | 10.30 |
ENSMUST00000239029.2
ENSMUST00000138395.9 ENSMUST00000156746.3 |
Ermap
|
erythroblast membrane-associated protein |
| chr4_-_133601990 | 9.75 |
ENSMUST00000168974.9
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr8_+_23629080 | 8.92 |
ENSMUST00000033947.15
|
Ank1
|
ankyrin 1, erythroid |
| chr7_+_99184858 | 8.86 |
ENSMUST00000032995.15
ENSMUST00000162404.8 |
Arrb1
|
arrestin, beta 1 |
| chr14_-_56322654 | 8.77 |
ENSMUST00000015594.9
|
Mcpt8
|
mast cell protease 8 |
| chr11_+_87684548 | 8.25 |
ENSMUST00000143021.9
|
Mpo
|
myeloperoxidase |
| chr4_-_119047167 | 8.24 |
ENSMUST00000030396.15
|
Ermap
|
erythroblast membrane-associated protein |
| chrX_+_8137881 | 8.03 |
ENSMUST00000115590.2
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr9_-_21874802 | 7.92 |
ENSMUST00000006397.7
|
Epor
|
erythropoietin receptor |
| chr8_+_85428059 | 7.87 |
ENSMUST00000238364.2
ENSMUST00000238562.2 ENSMUST00000037165.6 |
Lyl1
|
lymphoblastomic leukemia 1 |
| chr7_-_142209755 | 7.49 |
ENSMUST00000178921.2
|
Igf2
|
insulin-like growth factor 2 |
| chr14_-_70873385 | 7.25 |
ENSMUST00000228295.2
ENSMUST00000022695.16 |
Dmtn
|
dematin actin binding protein |
| chr17_-_35304582 | 7.16 |
ENSMUST00000038507.7
|
Ly6g6f
|
lymphocyte antigen 6 complex, locus G6F |
| chr1_-_170755136 | 7.11 |
ENSMUST00000046322.14
ENSMUST00000159171.2 |
Fcrla
|
Fc receptor-like A |
| chr2_+_164790139 | 7.10 |
ENSMUST00000017881.3
|
Mmp9
|
matrix metallopeptidase 9 |
| chr19_-_5776268 | 7.10 |
ENSMUST00000075606.6
ENSMUST00000236215.2 ENSMUST00000235730.2 ENSMUST00000237081.2 ENSMUST00000049295.15 |
Ehbp1l1
|
EH domain binding protein 1-like 1 |
| chr16_-_16687119 | 6.77 |
ENSMUST00000075017.5
|
Vpreb1
|
pre-B lymphocyte gene 1 |
| chr17_+_36132567 | 6.74 |
ENSMUST00000003635.7
|
Ier3
|
immediate early response 3 |
| chr11_+_104468107 | 6.63 |
ENSMUST00000106956.10
|
Myl4
|
myosin, light polypeptide 4 |
| chr9_-_44253588 | 6.62 |
ENSMUST00000215091.2
|
Hmbs
|
hydroxymethylbilane synthase |
| chr8_+_94905710 | 6.61 |
ENSMUST00000034215.8
ENSMUST00000212291.2 ENSMUST00000211807.2 |
Mt1
|
metallothionein 1 |
| chr16_+_18247666 | 6.49 |
ENSMUST00000144233.3
|
Txnrd2
|
thioredoxin reductase 2 |
| chr8_+_23629046 | 6.32 |
ENSMUST00000121075.8
|
Ank1
|
ankyrin 1, erythroid |
| chr4_-_119047146 | 6.29 |
ENSMUST00000124626.9
|
Ermap
|
erythroblast membrane-associated protein |
| chr4_-_119047180 | 6.28 |
ENSMUST00000150864.3
ENSMUST00000141227.9 |
Ermap
|
erythroblast membrane-associated protein |
| chr14_-_44112974 | 6.22 |
ENSMUST00000179200.2
|
Ear1
|
eosinophil-associated, ribonuclease A family, member 1 |
| chr8_+_73488496 | 6.14 |
ENSMUST00000058099.9
|
F2rl3
|
coagulation factor II (thrombin) receptor-like 3 |
| chr11_+_69737437 | 5.98 |
ENSMUST00000152566.8
ENSMUST00000108633.9 |
Plscr3
|
phospholipid scramblase 3 |
| chr19_+_58717319 | 5.93 |
ENSMUST00000048644.6
ENSMUST00000236445.2 |
Pnliprp1
|
pancreatic lipase related protein 1 |
| chr17_+_25517363 | 5.89 |
ENSMUST00000037453.4
|
Prss34
|
protease, serine 34 |
| chr11_+_117740077 | 5.84 |
ENSMUST00000081387.11
|
Birc5
|
baculoviral IAP repeat-containing 5 |
| chr1_+_63215976 | 5.82 |
ENSMUST00000129339.8
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr6_+_145067457 | 5.78 |
ENSMUST00000032396.13
|
Lrmp
|
lymphoid-restricted membrane protein |
| chr5_-_31102829 | 5.77 |
ENSMUST00000031051.8
|
Cgref1
|
cell growth regulator with EF hand domain 1 |
| chr6_+_72074545 | 5.75 |
ENSMUST00000069994.11
ENSMUST00000114112.4 |
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr11_+_69737491 | 5.74 |
ENSMUST00000019605.4
|
Plscr3
|
phospholipid scramblase 3 |
| chr6_+_66512401 | 5.74 |
ENSMUST00000101343.2
|
Mad2l1
|
MAD2 mitotic arrest deficient-like 1 |
| chr11_+_69737200 | 5.68 |
ENSMUST00000108632.8
|
Plscr3
|
phospholipid scramblase 3 |
| chr19_+_4644425 | 5.67 |
ENSMUST00000238089.2
|
Pcx
|
pyruvate carboxylase |
| chr11_-_69838971 | 5.55 |
ENSMUST00000179298.3
ENSMUST00000018710.13 ENSMUST00000135437.3 ENSMUST00000141837.9 ENSMUST00000142500.8 |
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr4_+_140427799 | 5.46 |
ENSMUST00000071169.9
|
Rcc2
|
regulator of chromosome condensation 2 |
| chr7_+_28140352 | 5.44 |
ENSMUST00000078845.13
|
Gmfg
|
glia maturation factor, gamma |
| chr14_-_63654478 | 5.42 |
ENSMUST00000014597.5
|
Blk
|
B lymphoid kinase |
| chr19_-_46033353 | 5.32 |
ENSMUST00000026252.14
ENSMUST00000156585.9 ENSMUST00000185355.7 ENSMUST00000152946.8 |
Ldb1
|
LIM domain binding 1 |
| chr7_-_126303351 | 5.29 |
ENSMUST00000106364.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr1_+_135656885 | 5.25 |
ENSMUST00000027677.8
|
Csrp1
|
cysteine and glycine-rich protein 1 |
| chr15_-_103161237 | 5.07 |
ENSMUST00000154510.8
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr9_+_62765362 | 5.02 |
ENSMUST00000213643.2
ENSMUST00000034777.14 ENSMUST00000163820.3 ENSMUST00000215870.2 ENSMUST00000214633.2 ENSMUST00000215968.2 |
Calml4
|
calmodulin-like 4 |
| chr1_+_134890288 | 4.96 |
ENSMUST00000027687.8
|
Ube2t
|
ubiquitin-conjugating enzyme E2T |
| chr2_+_130119077 | 4.95 |
ENSMUST00000028890.15
ENSMUST00000159373.2 |
Nop56
|
NOP56 ribonucleoprotein |
| chr10_+_128744689 | 4.88 |
ENSMUST00000105229.9
|
Cd63
|
CD63 antigen |
| chr11_+_116422570 | 4.88 |
ENSMUST00000106387.9
|
Sphk1
|
sphingosine kinase 1 |
| chr11_+_116422712 | 4.85 |
ENSMUST00000100201.10
|
Sphk1
|
sphingosine kinase 1 |
| chr1_+_107456731 | 4.68 |
ENSMUST00000182198.8
|
Serpinb10
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr2_-_153079828 | 4.68 |
ENSMUST00000109795.2
|
Plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr5_-_148336711 | 4.62 |
ENSMUST00000048116.15
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr17_-_26160872 | 4.56 |
ENSMUST00000139226.2
ENSMUST00000097368.10 ENSMUST00000140304.2 ENSMUST00000026823.16 |
Pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr4_-_132073048 | 4.56 |
ENSMUST00000084250.11
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr10_+_79650496 | 4.54 |
ENSMUST00000218857.2
ENSMUST00000220365.2 |
Palm
|
paralemmin |
| chr4_-_41098174 | 4.41 |
ENSMUST00000055327.8
|
Aqp3
|
aquaporin 3 |
| chr4_-_132072988 | 4.40 |
ENSMUST00000030726.13
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr5_+_122347792 | 4.31 |
ENSMUST00000072602.14
|
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr15_-_89310060 | 4.25 |
ENSMUST00000109313.9
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr5_+_122348140 | 4.23 |
ENSMUST00000196187.5
ENSMUST00000100747.3 |
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr1_-_170755109 | 4.22 |
ENSMUST00000162136.2
ENSMUST00000162887.2 |
Fcrla
|
Fc receptor-like A |
| chr9_-_44253630 | 4.22 |
ENSMUST00000097558.5
|
Hmbs
|
hydroxymethylbilane synthase |
| chr7_+_28140450 | 4.19 |
ENSMUST00000135686.2
|
Gmfg
|
glia maturation factor, gamma |
| chr18_+_34758062 | 4.15 |
ENSMUST00000166044.3
|
Kif20a
|
kinesin family member 20A |
| chr5_+_30824121 | 4.10 |
ENSMUST00000144742.6
ENSMUST00000149759.2 ENSMUST00000199320.5 |
Cenpa
|
centromere protein A |
| chr12_+_83572774 | 4.08 |
ENSMUST00000223291.2
|
Dcaf4
|
DDB1 and CUL4 associated factor 4 |
| chr6_+_91492988 | 4.06 |
ENSMUST00000206947.2
|
Lsm3
|
LSM3 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr16_-_18880821 | 4.04 |
ENSMUST00000200568.2
|
Iglc1
|
immunoglobulin lambda constant 1 |
| chr9_-_44255456 | 4.02 |
ENSMUST00000077353.15
|
Hmbs
|
hydroxymethylbilane synthase |
| chr1_+_63216281 | 4.02 |
ENSMUST00000188524.2
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr6_+_91492910 | 4.00 |
ENSMUST00000040607.6
|
Lsm3
|
LSM3 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr7_-_126398343 | 3.97 |
ENSMUST00000032934.12
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr17_+_25235039 | 3.96 |
ENSMUST00000142000.9
ENSMUST00000137386.8 |
Ift140
|
intraflagellar transport 140 |
| chr6_-_70149254 | 3.94 |
ENSMUST00000197272.2
|
Igkv8-27
|
immunoglobulin kappa chain variable 8-27 |
| chr17_+_48607405 | 3.94 |
ENSMUST00000170941.3
|
Treml2
|
triggering receptor expressed on myeloid cells-like 2 |
| chr2_-_31973795 | 3.92 |
ENSMUST00000056406.7
|
Fam78a
|
family with sequence similarity 78, member A |
| chr14_+_66534478 | 3.92 |
ENSMUST00000022623.13
|
Trim35
|
tripartite motif-containing 35 |
| chr6_-_5496261 | 3.91 |
ENSMUST00000203347.3
ENSMUST00000019721.7 |
Pdk4
|
pyruvate dehydrogenase kinase, isoenzyme 4 |
| chr7_-_126398165 | 3.81 |
ENSMUST00000205890.2
ENSMUST00000205336.2 ENSMUST00000087566.11 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr10_+_75399920 | 3.80 |
ENSMUST00000141062.8
ENSMUST00000152657.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr10_+_127350820 | 3.76 |
ENSMUST00000035735.11
|
Ndufa4l2
|
Ndufa4, mitochondrial complex associated like 2 |
| chr15_+_78784043 | 3.76 |
ENSMUST00000001226.11
|
Sh3bp1
|
SH3-domain binding protein 1 |
| chr8_-_72175949 | 3.74 |
ENSMUST00000125092.2
|
Fcho1
|
FCH domain only 1 |
| chr19_-_4240984 | 3.71 |
ENSMUST00000045864.4
|
Tbc1d10c
|
TBC1 domain family, member 10c |
| chr6_+_66512374 | 3.70 |
ENSMUST00000116605.8
|
Mad2l1
|
MAD2 mitotic arrest deficient-like 1 |
| chr18_+_34757687 | 3.67 |
ENSMUST00000237407.2
|
Kif20a
|
kinesin family member 20A |
| chr12_+_31123860 | 3.66 |
ENSMUST00000041133.10
|
Fam110c
|
family with sequence similarity 110, member C |
| chr9_+_110173253 | 3.63 |
ENSMUST00000199709.3
|
Scap
|
SREBF chaperone |
| chr7_+_127846121 | 3.56 |
ENSMUST00000167965.8
|
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr10_-_60588539 | 3.56 |
ENSMUST00000117513.8
ENSMUST00000119595.8 |
Slc29a3
|
solute carrier family 29 (nucleoside transporters), member 3 |
| chr10_+_80692948 | 3.52 |
ENSMUST00000220091.2
|
Sppl2b
|
signal peptide peptidase like 2B |
| chr7_+_127845984 | 3.47 |
ENSMUST00000164710.8
ENSMUST00000070656.12 |
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr18_+_34757666 | 3.45 |
ENSMUST00000167161.9
|
Kif20a
|
kinesin family member 20A |
| chr16_+_17798292 | 3.44 |
ENSMUST00000075371.5
|
Vpreb2
|
pre-B lymphocyte gene 2 |
| chr15_+_79784365 | 3.43 |
ENSMUST00000230135.2
|
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr10_+_79833296 | 3.41 |
ENSMUST00000171637.8
ENSMUST00000043866.8 |
Abca7
|
ATP-binding cassette, sub-family A (ABC1), member 7 |
| chr4_-_155737841 | 3.38 |
ENSMUST00000030937.2
|
Mmp23
|
matrix metallopeptidase 23 |
| chr9_-_42035560 | 3.37 |
ENSMUST00000060989.9
|
Sorl1
|
sortilin-related receptor, LDLR class A repeats-containing |
| chr1_+_152683627 | 3.36 |
ENSMUST00000027754.7
|
Ncf2
|
neutrophil cytosolic factor 2 |
| chr16_-_19801781 | 3.35 |
ENSMUST00000058839.10
|
Klhl6
|
kelch-like 6 |
| chr7_+_46496929 | 3.28 |
ENSMUST00000132157.2
ENSMUST00000210631.2 |
Ldha
|
lactate dehydrogenase A |
| chr19_-_7688628 | 3.28 |
ENSMUST00000025666.8
|
Slc22a19
|
solute carrier family 22 (organic anion transporter), member 19 |
| chr12_-_110945415 | 3.27 |
ENSMUST00000135131.2
ENSMUST00000043459.13 ENSMUST00000128353.8 |
Ankrd9
|
ankyrin repeat domain 9 |
| chrX_-_9335525 | 3.25 |
ENSMUST00000015484.10
|
Cybb
|
cytochrome b-245, beta polypeptide |
| chr4_-_131802561 | 3.24 |
ENSMUST00000105970.8
ENSMUST00000105975.8 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr16_+_49675682 | 3.19 |
ENSMUST00000114496.3
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr7_+_125202653 | 3.19 |
ENSMUST00000206103.2
ENSMUST00000033000.8 |
Il21r
|
interleukin 21 receptor |
| chr4_-_49597425 | 3.19 |
ENSMUST00000150664.2
|
Pgap4
|
post-GPI attachment to proteins GalNAc transferase 4 |
| chr10_-_81335966 | 3.17 |
ENSMUST00000053646.7
|
S1pr4
|
sphingosine-1-phosphate receptor 4 |
| chr7_+_46496552 | 3.14 |
ENSMUST00000005051.6
|
Ldha
|
lactate dehydrogenase A |
| chr5_+_145282064 | 3.13 |
ENSMUST00000079268.9
|
Cyp3a57
|
cytochrome P450, family 3, subfamily a, polypeptide 57 |
| chr1_+_91468409 | 3.11 |
ENSMUST00000027538.9
ENSMUST00000190484.7 ENSMUST00000186068.2 |
Asb1
|
ankyrin repeat and SOCS box-containing 1 |
| chr4_-_131802606 | 3.11 |
ENSMUST00000146021.8
|
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr6_-_70051586 | 3.11 |
ENSMUST00000103377.3
|
Igkv6-32
|
immunoglobulin kappa variable 6-32 |
| chr8_+_85428391 | 3.10 |
ENSMUST00000238338.2
|
Lyl1
|
lymphoblastomic leukemia 1 |
| chr19_-_4109446 | 3.09 |
ENSMUST00000189808.7
|
Gstp3
|
glutathione S-transferase pi 3 |
| chr4_+_52439237 | 3.08 |
ENSMUST00000102915.10
ENSMUST00000117280.8 ENSMUST00000142227.3 |
Smc2
|
structural maintenance of chromosomes 2 |
| chr3_-_86049988 | 3.05 |
ENSMUST00000029722.7
|
Rps3a1
|
ribosomal protein S3A1 |
| chr4_+_132701413 | 3.05 |
ENSMUST00000030693.13
|
Fgr
|
FGR proto-oncogene, Src family tyrosine kinase |
| chr11_-_69786324 | 3.04 |
ENSMUST00000001631.7
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr8_-_22675773 | 3.00 |
ENSMUST00000046916.9
|
Ckap2
|
cytoskeleton associated protein 2 |
| chr3_+_157272504 | 2.98 |
ENSMUST00000041175.13
ENSMUST00000173533.2 |
Ptger3
|
prostaglandin E receptor 3 (subtype EP3) |
| chr2_+_91376650 | 2.96 |
ENSMUST00000099716.11
ENSMUST00000046769.16 ENSMUST00000111337.3 |
Ckap5
|
cytoskeleton associated protein 5 |
| chr2_+_163500290 | 2.95 |
ENSMUST00000164399.8
ENSMUST00000064703.13 ENSMUST00000099105.9 ENSMUST00000152418.8 ENSMUST00000126182.8 ENSMUST00000131228.8 |
Pkig
|
protein kinase inhibitor, gamma |
| chr3_+_51131868 | 2.94 |
ENSMUST00000023849.15
ENSMUST00000167780.2 |
Noct
|
nocturnin |
| chr6_-_124710030 | 2.93 |
ENSMUST00000173647.2
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr7_+_141995545 | 2.93 |
ENSMUST00000105971.8
ENSMUST00000145287.8 |
Tnni2
|
troponin I, skeletal, fast 2 |
| chr19_-_4241034 | 2.93 |
ENSMUST00000237495.2
|
Tbc1d10c
|
TBC1 domain family, member 10c |
| chr2_+_163503415 | 2.92 |
ENSMUST00000135537.8
|
Pkig
|
protein kinase inhibitor, gamma |
| chr17_-_33937565 | 2.91 |
ENSMUST00000174040.2
ENSMUST00000173015.8 ENSMUST00000066121.13 ENSMUST00000186022.7 ENSMUST00000173329.8 ENSMUST00000172767.9 |
Marchf2
|
membrane associated ring-CH-type finger 2 |
| chr10_+_117465397 | 2.91 |
ENSMUST00000020399.6
|
Cpm
|
carboxypeptidase M |
| chr11_+_113510135 | 2.90 |
ENSMUST00000146390.3
|
Sstr2
|
somatostatin receptor 2 |
| chr5_-_33432310 | 2.86 |
ENSMUST00000201372.3
ENSMUST00000202962.4 ENSMUST00000201575.4 ENSMUST00000202868.4 ENSMUST00000079746.10 |
Ctbp1
|
C-terminal binding protein 1 |
| chr7_+_89814713 | 2.84 |
ENSMUST00000207084.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr6_+_125529911 | 2.79 |
ENSMUST00000112254.8
ENSMUST00000112253.6 |
Vwf
|
Von Willebrand factor |
| chr10_+_81012465 | 2.78 |
ENSMUST00000047864.11
|
Eef2
|
eukaryotic translation elongation factor 2 |
| chr3_-_59118293 | 2.78 |
ENSMUST00000040622.3
|
P2ry13
|
purinergic receptor P2Y, G-protein coupled 13 |
| chr15_+_80507671 | 2.76 |
ENSMUST00000043149.9
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr7_-_125090757 | 2.76 |
ENSMUST00000033006.14
|
Nsmce1
|
NSE1 homolog, SMC5-SMC6 complex component |
| chr12_-_114443071 | 2.72 |
ENSMUST00000103492.2
|
Ighv10-1
|
immunoglobulin heavy variable 10-1 |
| chr16_+_49675969 | 2.67 |
ENSMUST00000229101.2
ENSMUST00000230836.2 ENSMUST00000229206.2 ENSMUST00000084838.14 ENSMUST00000230281.2 |
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr7_+_110376859 | 2.67 |
ENSMUST00000148292.2
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr7_-_24919247 | 2.62 |
ENSMUST00000058702.7
|
Dedd2
|
death effector domain-containing DNA binding protein 2 |
| chr12_-_69837434 | 2.62 |
ENSMUST00000021377.5
|
Cdkl1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
| chr14_+_24540745 | 2.61 |
ENSMUST00000112384.10
|
Rps24
|
ribosomal protein S24 |
| chr14_-_30329765 | 2.61 |
ENSMUST00000112207.8
ENSMUST00000112206.8 ENSMUST00000112202.8 ENSMUST00000112203.2 |
Prkcd
|
protein kinase C, delta |
| chr10_-_75600100 | 2.60 |
ENSMUST00000218469.2
ENSMUST00000001712.8 |
Cabin1
|
calcineurin binding protein 1 |
| chr2_+_71219561 | 2.59 |
ENSMUST00000028408.3
|
Hat1
|
histone aminotransferase 1 |
| chr6_-_69584812 | 2.59 |
ENSMUST00000103359.3
|
Igkv4-55
|
immunoglobulin kappa variable 4-55 |
| chr15_+_78783867 | 2.58 |
ENSMUST00000134703.8
ENSMUST00000061239.14 ENSMUST00000109698.9 |
Gm49510
Sh3bp1
|
predicted gene, 49510 SH3-domain binding protein 1 |
| chr15_-_86070338 | 2.58 |
ENSMUST00000044332.16
|
Cerk
|
ceramide kinase |
| chr7_+_46496506 | 2.57 |
ENSMUST00000209984.2
|
Ldha
|
lactate dehydrogenase A |
| chr14_+_24540815 | 2.55 |
ENSMUST00000224568.2
|
Rps24
|
ribosomal protein S24 |
| chr9_+_45314436 | 2.52 |
ENSMUST00000041005.6
|
Fxyd2
|
FXYD domain-containing ion transport regulator 2 |
| chr15_-_102097387 | 2.49 |
ENSMUST00000230288.2
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr12_+_69215583 | 2.46 |
ENSMUST00000110621.3
ENSMUST00000222520.2 |
Lrr1
|
leucine rich repeat protein 1 |
| chr6_-_143045731 | 2.45 |
ENSMUST00000203673.3
ENSMUST00000203187.3 ENSMUST00000171349.8 ENSMUST00000087485.7 |
C2cd5
|
C2 calcium-dependent domain containing 5 |
| chr11_-_121120052 | 2.44 |
ENSMUST00000169393.8
ENSMUST00000106115.8 ENSMUST00000038709.14 ENSMUST00000147490.6 |
Cybc1
|
cytochrome b 245 chaperone 1 |
| chr12_+_113112311 | 2.42 |
ENSMUST00000199089.5
|
Crip1
|
cysteine-rich protein 1 (intestinal) |
| chr16_-_20245071 | 2.42 |
ENSMUST00000115547.9
ENSMUST00000096199.5 |
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr19_+_10819896 | 2.41 |
ENSMUST00000025646.3
|
Slc15a3
|
solute carrier family 15, member 3 |
| chr2_-_180928867 | 2.40 |
ENSMUST00000130475.8
|
Gmeb2
|
glucocorticoid modulatory element binding protein 2 |
| chr5_+_145051025 | 2.40 |
ENSMUST00000085679.13
|
Arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr3_+_88744323 | 2.40 |
ENSMUST00000081695.14
ENSMUST00000090942.6 |
Gon4l
|
gon-4-like (C.elegans) |
| chr2_+_154633265 | 2.38 |
ENSMUST00000140713.3
ENSMUST00000137333.8 |
Raly
a
|
hnRNP-associated with lethal yellow nonagouti |
| chr4_-_59783780 | 2.38 |
ENSMUST00000107526.8
ENSMUST00000095063.11 |
Inip
|
INTS3 and NABP interacting protein |
| chr14_+_62569517 | 2.38 |
ENSMUST00000022499.13
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chr5_-_113957318 | 2.37 |
ENSMUST00000201194.4
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr11_-_59054521 | 2.37 |
ENSMUST00000137433.2
ENSMUST00000054523.6 |
Iba57
|
IBA57 homolog, iron-sulfur cluster assembly |
| chr6_+_4504814 | 2.37 |
ENSMUST00000141483.8
|
Col1a2
|
collagen, type I, alpha 2 |
| chr14_+_24540777 | 2.37 |
ENSMUST00000169826.3
ENSMUST00000225023.2 ENSMUST00000223999.2 |
Rps24
|
ribosomal protein S24 |
| chr17_-_79662514 | 2.35 |
ENSMUST00000068958.9
|
Cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr6_-_124710084 | 2.34 |
ENSMUST00000112484.10
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr5_-_151574620 | 2.34 |
ENSMUST00000038131.10
|
Rfc3
|
replication factor C (activator 1) 3 |
| chr13_+_69950509 | 2.32 |
ENSMUST00000223376.2
ENSMUST00000222387.2 |
Med10
|
mediator complex subunit 10 |
| chr4_-_149783097 | 2.32 |
ENSMUST00000038859.14
ENSMUST00000105690.9 |
Pik3cd
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta |
| chr13_-_107073415 | 2.28 |
ENSMUST00000080856.14
|
Ipo11
|
importin 11 |
| chr5_+_122347912 | 2.26 |
ENSMUST00000143560.8
|
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr7_-_30741497 | 2.25 |
ENSMUST00000162116.8
ENSMUST00000159924.8 |
Fxyd5
|
FXYD domain-containing ion transport regulator 5 |
| chr15_+_79982033 | 2.25 |
ENSMUST00000143928.2
|
Syngr1
|
synaptogyrin 1 |
| chr9_-_66032134 | 2.23 |
ENSMUST00000034946.15
|
Snx1
|
sorting nexin 1 |
| chr14_+_78141679 | 2.22 |
ENSMUST00000022591.16
ENSMUST00000169978.2 ENSMUST00000227903.2 |
Epsti1
|
epithelial stromal interaction 1 (breast) |
| chr11_-_5015275 | 2.22 |
ENSMUST00000109895.2
ENSMUST00000152257.2 ENSMUST00000037146.10 ENSMUST00000056649.13 |
Gas2l1
|
growth arrest-specific 2 like 1 |
| chr18_-_36648850 | 2.19 |
ENSMUST00000025363.7
|
Hbegf
|
heparin-binding EGF-like growth factor |
| chr11_+_51510555 | 2.14 |
ENSMUST00000127405.2
|
Nhp2
|
NHP2 ribonucleoprotein |
| chrX_-_165992311 | 2.12 |
ENSMUST00000112172.4
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
Network of associatons between targets according to the STRING database.
First level regulatory network of Esr1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.0 | 42.0 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 6.4 | 19.1 | GO:0002780 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) neutrophil mediated killing of fungus(GO:0070947) |
| 5.3 | 21.2 | GO:0044010 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 5.0 | 14.9 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 3.2 | 9.6 | GO:0071846 | actin filament debranching(GO:0071846) |
| 2.4 | 7.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 1.8 | 5.5 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 1.8 | 5.4 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 1.8 | 5.3 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 1.6 | 4.7 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 1.5 | 29.0 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 1.4 | 9.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 1.2 | 7.2 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 1.2 | 5.9 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 1.2 | 17.4 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 1.1 | 5.7 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 1.1 | 5.4 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 1.1 | 9.8 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 1.1 | 8.7 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 1.1 | 8.5 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 1.0 | 9.4 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 1.0 | 10.0 | GO:0019661 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 1.0 | 3.0 | GO:1904328 | gastrin-induced gastric acid secretion(GO:0001698) positive regulation of actin filament-based movement(GO:1903116) regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 1.0 | 5.0 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 1.0 | 3.9 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 1.0 | 2.9 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.9 | 2.8 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.9 | 12.0 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.9 | 11.0 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.9 | 2.7 | GO:0006233 | dTDP biosynthetic process(GO:0006233) dTDP metabolic process(GO:0046072) |
| 0.8 | 5.8 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.8 | 4.9 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.8 | 4.5 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.7 | 6.7 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.7 | 6.6 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.7 | 2.2 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 0.7 | 18.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.7 | 5.3 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.7 | 2.6 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.6 | 5.2 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.6 | 1.9 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.6 | 8.0 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.6 | 1.8 | GO:2000977 | regulation of forebrain neuron differentiation(GO:2000977) |
| 0.6 | 2.4 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.6 | 1.2 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) positive regulation of transforming growth factor beta3 production(GO:0032916) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.6 | 2.8 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.6 | 6.2 | GO:0008228 | opsonization(GO:0008228) |
| 0.5 | 3.3 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.5 | 5.4 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.5 | 1.6 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.5 | 11.1 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.5 | 2.1 | GO:0021941 | negative regulation of cerebellar granule cell precursor proliferation(GO:0021941) |
| 0.5 | 1.6 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.5 | 1.5 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.5 | 2.1 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.5 | 5.0 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.5 | 3.0 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.5 | 5.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.5 | 11.3 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.5 | 1.5 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.5 | 3.8 | GO:0031179 | peptide modification(GO:0031179) |
| 0.5 | 1.9 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) negative regulation of t-circle formation(GO:1904430) |
| 0.5 | 2.4 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.5 | 7.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.4 | 4.0 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.4 | 8.0 | GO:0015816 | glycine transport(GO:0015816) |
| 0.4 | 1.3 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.4 | 1.3 | GO:0014739 | positive regulation of muscle hyperplasia(GO:0014739) |
| 0.4 | 1.3 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.4 | 2.8 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.4 | 1.2 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.4 | 3.5 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.4 | 1.2 | GO:0036145 | dendritic cell homeostasis(GO:0036145) |
| 0.4 | 1.9 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.4 | 1.8 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.4 | 2.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.4 | 1.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.4 | 2.5 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.4 | 6.4 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.3 | 1.0 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.3 | 3.1 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.3 | 5.1 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.3 | 11.0 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.3 | 4.9 | GO:0030432 | peristalsis(GO:0030432) |
| 0.3 | 12.9 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.3 | 1.5 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.3 | 1.2 | GO:0009816 | defense response, incompatible interaction(GO:0009814) defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.3 | 0.9 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.3 | 1.1 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.3 | 1.4 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.3 | 1.4 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.3 | 5.9 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.3 | 10.8 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.3 | 0.8 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.3 | 4.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.3 | 1.4 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.3 | 1.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.3 | 2.7 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.3 | 1.1 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.3 | 4.0 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.3 | 1.6 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.3 | 2.3 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.3 | 4.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.3 | 4.1 | GO:0071459 | protein localization to chromosome, centromeric region(GO:0071459) |
| 0.3 | 1.3 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.3 | 1.0 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.3 | 4.3 | GO:0015809 | arginine transport(GO:0015809) |
| 0.3 | 2.0 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.2 | 0.7 | GO:1904632 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) response to diterpene(GO:1904629) cellular response to diterpene(GO:1904630) response to glucoside(GO:1904631) cellular response to glucoside(GO:1904632) |
| 0.2 | 0.7 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.2 | 2.6 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 0.5 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.2 | 2.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.2 | 6.2 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.2 | 0.9 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 0.2 | 0.7 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.2 | 1.1 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.2 | 2.1 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.2 | 0.6 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.2 | 1.6 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.2 | 0.4 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.2 | 1.7 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.2 | 0.6 | GO:1903632 | positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) |
| 0.2 | 0.8 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.2 | 2.1 | GO:0035878 | nail development(GO:0035878) |
| 0.2 | 1.7 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.2 | 4.8 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.2 | 0.6 | GO:0070476 | rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.2 | 8.0 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.2 | 0.9 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.2 | 0.5 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.2 | 4.0 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.2 | 0.9 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.2 | 2.0 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.2 | 2.1 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.2 | 2.6 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.2 | 1.5 | GO:0071231 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.2 | 2.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.2 | 1.7 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.2 | 3.7 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.2 | 1.3 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.2 | 2.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.2 | 10.1 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.2 | 0.3 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.2 | 1.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.2 | 2.4 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.2 | 3.6 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.2 | 4.6 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.2 | 2.4 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 | 0.5 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.2 | 0.8 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.2 | 2.4 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.1 | 9.2 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 0.6 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.1 | 6.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 2.8 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.1 | 2.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 1.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.7 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.1 | 0.8 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 1.0 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.1 | 0.5 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 2.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.5 | GO:0051311 | meiotic metaphase I plate congression(GO:0043060) meiotic metaphase plate congression(GO:0051311) |
| 0.1 | 0.4 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.1 | 0.7 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.1 | 3.5 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.1 | 1.2 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 5.1 | GO:0000305 | response to oxygen radical(GO:0000305) |
| 0.1 | 0.7 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.1 | 3.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 5.9 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.1 | 2.4 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 | 0.9 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.1 | 1.8 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 2.0 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 1.0 | GO:1905146 | lysosomal lumen acidification(GO:0007042) lysosomal protein catabolic process(GO:1905146) |
| 0.1 | 1.0 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.1 | 2.4 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 3.8 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.1 | 0.4 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.1 | 24.8 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 1.5 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.1 | 2.4 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.1 | 3.0 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 | 15.9 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 0.7 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 0.3 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.1 | 0.4 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.1 | 1.2 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.1 | 0.6 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.1 | 4.0 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 1.4 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 0.4 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 0.7 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 0.8 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 2.1 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 0.3 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 1.1 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.1 | 0.4 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 3.3 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.1 | 0.5 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 1.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 1.4 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.1 | 0.2 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.1 | 4.2 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.1 | 0.8 | GO:0031077 | post-embryonic camera-type eye development(GO:0031077) |
| 0.1 | 4.3 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 0.8 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 1.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 0.4 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 0.8 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 1.7 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 1.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 5.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 3.2 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 0.3 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.1 | 1.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 10.4 | GO:0071222 | cellular response to lipopolysaccharide(GO:0071222) |
| 0.1 | 1.0 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 0.3 | GO:0006222 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.1 | 2.9 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 0.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.3 | GO:0060082 | response to carbon monoxide(GO:0034465) eye blink reflex(GO:0060082) |
| 0.1 | 0.5 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.1 | 0.3 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.1 | 0.3 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 1.9 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 1.9 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 1.2 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.6 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.2 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.0 | 0.5 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 1.1 | GO:0030262 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 1.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 1.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.7 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.5 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.6 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 1.2 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.7 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 1.5 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 2.0 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.0 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 1.7 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.5 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.5 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.0 | 1.3 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.0 | 0.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 1.0 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 1.1 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.6 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 1.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 1.3 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.6 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 1.2 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.0 | 0.1 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) RNA surveillance(GO:0071025) |
| 0.0 | 1.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 1.1 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 0.5 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.3 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 1.5 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 1.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.3 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 1.2 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.8 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.0 | 0.1 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 1.5 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 1.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 1.6 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.7 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.2 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.0 | 0.0 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.4 | GO:0090662 | ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 1.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 4.3 | GO:0008380 | RNA splicing(GO:0008380) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 21.2 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 2.4 | 41.3 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 1.6 | 4.9 | GO:0031904 | endosome lumen(GO:0031904) |
| 1.5 | 5.8 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 1.4 | 9.8 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 1.0 | 7.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 1.0 | 4.1 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.9 | 8.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.8 | 2.4 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.7 | 3.0 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.7 | 2.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.7 | 17.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.7 | 2.0 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.6 | 5.8 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.6 | 14.0 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.6 | 2.4 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.6 | 3.5 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.6 | 8.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.5 | 2.1 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.4 | 9.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.4 | 5.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.4 | 5.0 | GO:0000796 | condensin complex(GO:0000796) |
| 0.4 | 2.8 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.4 | 4.0 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.4 | 2.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.4 | 2.4 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.4 | 4.0 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.4 | 2.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.4 | 1.4 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.3 | 4.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.3 | 2.0 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.3 | 1.6 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.3 | 10.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.3 | 5.6 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.3 | 2.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.3 | 2.8 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.3 | 2.8 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.3 | 1.6 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.3 | 1.5 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.3 | 4.6 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.2 | 4.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 1.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.2 | 5.5 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.2 | 0.7 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.2 | 1.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 6.6 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.2 | 2.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.2 | 2.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 1.5 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.2 | 1.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.2 | 4.9 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 0.5 | GO:0005715 | chiasma(GO:0005712) late recombination nodule(GO:0005715) |
| 0.2 | 0.6 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.2 | 2.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 1.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 1.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) endoplasmic reticulum Sec complex(GO:0031205) translocon complex(GO:0071256) |
| 0.2 | 9.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 2.4 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.6 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 17.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 8.7 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 3.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 11.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.0 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 20.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.8 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 1.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.8 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 4.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 10.0 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 0.3 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 3.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 3.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.5 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.5 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 2.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 22.4 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.1 | 1.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 1.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 1.7 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 1.1 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 1.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.4 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 3.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.6 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.7 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 2.1 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.4 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 8.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 3.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 1.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 1.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.7 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 1.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.4 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.1 | 1.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 3.4 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 0.7 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 8.6 | GO:0005819 | spindle(GO:0005819) |
| 0.1 | 2.1 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.0 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.4 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 9.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 5.4 | GO:0042571 | immunoglobulin complex(GO:0019814) immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 1.7 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 2.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 3.8 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.7 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 2.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.8 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 3.5 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 3.7 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 1.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 2.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 1.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 4.0 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.5 | GO:0032155 | cell division site(GO:0032153) cleavage furrow(GO:0032154) cell division site part(GO:0032155) cell surface furrow(GO:0097610) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.9 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 9.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.2 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.7 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.2 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 14.9 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 3.0 | 8.9 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 2.0 | 8.1 | GO:0030629 | U6 snRNA 3'-end binding(GO:0030629) |
| 1.9 | 5.7 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 1.8 | 10.8 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 1.6 | 15.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 1.5 | 6.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 1.4 | 5.7 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 1.3 | 9.0 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 1.2 | 7.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 1.1 | 3.4 | GO:0034188 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 1.1 | 4.5 | GO:0070976 | TIR domain binding(GO:0070976) |
| 1.1 | 5.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 1.0 | 32.1 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 1.0 | 8.8 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 1.0 | 3.9 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.9 | 10.0 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.9 | 2.7 | GO:0004798 | thymidylate kinase activity(GO:0004798) |
| 0.9 | 19.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.9 | 4.4 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.9 | 2.6 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.8 | 5.9 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.8 | 8.7 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.8 | 7.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.7 | 4.4 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.7 | 2.1 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 0.7 | 4.2 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.7 | 21.6 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.7 | 9.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.6 | 4.5 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.6 | 5.8 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.6 | 5.9 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.6 | 17.5 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.6 | 8.0 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.6 | 2.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.5 | 5.4 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.5 | 1.6 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.5 | 3.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.5 | 3.0 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.5 | 12.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.5 | 2.0 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.5 | 1.9 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.5 | 2.8 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.5 | 1.9 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.4 | 3.0 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.4 | 28.9 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.4 | 5.3 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.4 | 20.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.4 | 3.1 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.4 | 1.9 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.4 | 1.9 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.4 | 1.8 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.4 | 6.5 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.4 | 1.4 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.4 | 5.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.3 | 1.7 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.3 | 2.4 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.3 | 4.6 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.3 | 16.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 5.7 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.3 | 15.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.3 | 4.0 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.3 | 1.5 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.3 | 2.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.3 | 3.7 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.3 | 2.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.3 | 2.9 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.3 | 9.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.3 | 1.7 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.3 | 0.8 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.3 | 0.8 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.3 | 2.7 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 1.5 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.3 | 1.3 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.2 | 1.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.2 | 5.9 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.2 | 1.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.2 | 1.5 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.2 | 1.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.2 | 11.9 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.2 | 2.0 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.2 | 1.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 2.3 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 3.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 1.4 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.2 | 7.1 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.2 | 1.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 1.7 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.2 | 2.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 7.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 2.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 0.5 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.2 | 3.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.2 | 0.9 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.2 | 2.6 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.2 | 0.7 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.2 | 0.7 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 9.9 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.2 | 3.1 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.2 | 0.8 | GO:0034618 | arginine-tRNA ligase activity(GO:0004814) arginine binding(GO:0034618) |
| 0.2 | 0.5 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.2 | 0.9 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 2.8 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 1.5 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.1 | 2.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 1.1 | GO:0052851 | ferric-chelate reductase activity(GO:0000293) cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.7 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.1 | 1.9 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 2.9 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 2.9 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 2.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.4 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.5 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 3.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.7 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.5 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.1 | 0.4 | GO:0070336 | forked DNA-dependent helicase activity(GO:0061749) flap-structured DNA binding(GO:0070336) |
| 0.1 | 1.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 0.7 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.5 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.1 | 1.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 4.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 1.9 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.3 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 0.5 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 11.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.6 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 5.1 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.1 | 2.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.7 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 0.1 | 0.7 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 2.8 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 2.6 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 1.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.8 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 6.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.9 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 1.3 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 3.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 4.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 1.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 0.5 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 0.9 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 1.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 9.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.4 | GO:0004905 | interferon receptor activity(GO:0004904) type I interferon receptor activity(GO:0004905) type I interferon binding(GO:0019962) |
| 0.1 | 5.9 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 3.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 5.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 1.9 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.8 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.9 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.1 | 0.5 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.7 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.7 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 3.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 0.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 1.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.4 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.1 | 1.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.8 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 4.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 0.2 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.7 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 1.6 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 3.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.0 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 2.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.0 | 0.3 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.3 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.4 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 1.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 1.2 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:1902121 | NADP+ binding(GO:0070401) lithocholic acid binding(GO:1902121) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.4 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 1.7 | GO:0016860 | intramolecular oxidoreductase activity(GO:0016860) |
| 0.0 | 1.0 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 4.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 3.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 1.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 10.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 1.7 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 1.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.0 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 1.5 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 1.9 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.0 | GO:0098634 | protein binding involved in cell-matrix adhesion(GO:0098634) integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.0 | 2.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 20.7 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 2.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.0 | 0.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.2 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.0 | 0.9 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 2.3 | GO:0005549 | odorant binding(GO:0005549) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 42.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.5 | 26.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.4 | 12.0 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.4 | 19.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.4 | 13.8 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.4 | 13.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.3 | 7.9 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.3 | 12.6 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.3 | 11.4 | PID EPO PATHWAY | EPO signaling pathway |
| 0.3 | 2.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 6.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.2 | 20.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 5.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 4.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.2 | 10.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 12.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.2 | 7.3 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 2.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 5.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 4.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 3.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 2.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 2.6 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 2.4 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 2.7 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 25.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 9.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 4.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 0.8 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 3.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 7.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 3.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 2.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.2 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 2.8 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.5 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 1.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.5 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.9 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.7 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.9 | PID NOTCH PATHWAY | Notch signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 26.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.6 | 14.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.6 | 29.8 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.6 | 13.9 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.5 | 10.9 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.4 | 7.0 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.4 | 7.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.4 | 9.4 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.4 | 8.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.4 | 1.5 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.4 | 11.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.3 | 11.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.3 | 17.2 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.3 | 4.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 3.0 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.3 | 15.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.3 | 13.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 4.5 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.2 | 15.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 9.4 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 3.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 18.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 2.9 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.2 | 4.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 5.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.2 | 2.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 10.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 7.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 14.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 1.1 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.2 | 2.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 9.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 4.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 2.6 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 2.4 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 5.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 12.3 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.1 | 2.1 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 5.2 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 2.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.8 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 4.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 1.1 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 0.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 0.9 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 2.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 0.9 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 5.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 4.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 0.4 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.1 | 1.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 3.6 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 2.5 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 1.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.0 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 1.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 4.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 3.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 2.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 4.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.7 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 1.0 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 4.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.3 | REACTOME PI3K EVENTS IN ERBB4 SIGNALING | Genes involved in PI3K events in ERBB4 signaling |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 4.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.3 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |