Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
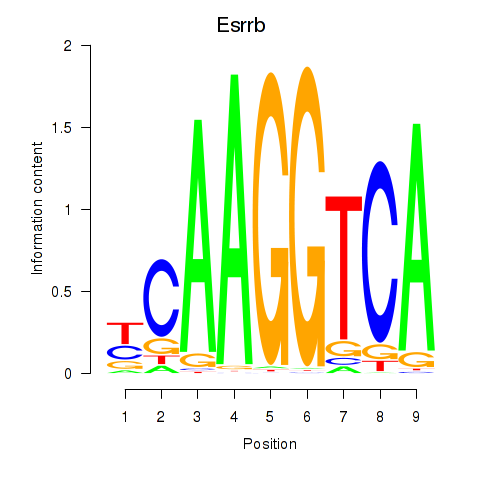
Results for Esrrb_Esrra
Z-value: 1.11
Transcription factors associated with Esrrb_Esrra
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Esrrb
|
ENSMUSG00000021255.18 | estrogen related receptor, beta |
|
Esrra
|
ENSMUSG00000024955.16 | estrogen related receptor, alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Esrra | mm39_v1_chr19_-_6899173_6899208 | 0.67 | 6.4e-06 | Click! |
| Esrrb | mm39_v1_chr12_+_86516875_86516897 | -0.31 | 7.0e-02 | Click! |
Activity profile of Esrrb_Esrra motif
Sorted Z-values of Esrrb_Esrra motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_43512929 | 18.30 |
ENSMUST00000026196.14
|
Got1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr15_+_76579885 | 11.93 |
ENSMUST00000231028.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr17_-_35077089 | 10.57 |
ENSMUST00000153400.8
|
Cfb
|
complement factor B |
| chr15_+_76579960 | 10.35 |
ENSMUST00000229679.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr2_+_102536701 | 9.42 |
ENSMUST00000123759.8
ENSMUST00000005220.11 ENSMUST00000111212.8 |
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr4_-_107164347 | 8.66 |
ENSMUST00000082426.11
|
Dio1
|
deiodinase, iodothyronine, type I |
| chr4_-_107164315 | 8.24 |
ENSMUST00000126291.2
ENSMUST00000106748.2 ENSMUST00000129138.2 |
Dio1
|
deiodinase, iodothyronine, type I |
| chr10_+_128030315 | 7.90 |
ENSMUST00000044776.13
|
Gls2
|
glutaminase 2 (liver, mitochondrial) |
| chr6_+_90527762 | 6.70 |
ENSMUST00000130418.8
ENSMUST00000032175.11 ENSMUST00000203111.2 |
Aldh1l1
|
aldehyde dehydrogenase 1 family, member L1 |
| chr18_+_45402018 | 6.39 |
ENSMUST00000183850.8
ENSMUST00000066890.14 |
Kcnn2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
| chr10_+_128030500 | 6.28 |
ENSMUST00000123291.2
|
Gls2
|
glutaminase 2 (liver, mitochondrial) |
| chr4_+_134123631 | 6.08 |
ENSMUST00000105869.9
|
Pafah2
|
platelet-activating factor acetylhydrolase 2 |
| chr7_-_105249308 | 5.59 |
ENSMUST00000210531.2
ENSMUST00000033185.10 |
Hpx
|
hemopexin |
| chr9_-_46146558 | 5.20 |
ENSMUST00000121916.8
ENSMUST00000034586.9 |
Apoc3
|
apolipoprotein C-III |
| chr9_-_46146928 | 4.69 |
ENSMUST00000118649.8
|
Apoc3
|
apolipoprotein C-III |
| chr15_+_25843225 | 4.62 |
ENSMUST00000022881.15
|
Retreg1
|
reticulophagy regulator 1 |
| chr15_+_7159038 | 4.19 |
ENSMUST00000067190.12
ENSMUST00000164529.9 |
Lifr
|
LIF receptor alpha |
| chr4_+_140688514 | 3.98 |
ENSMUST00000010007.9
|
Sdhb
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr17_-_32639936 | 3.95 |
ENSMUST00000170392.9
ENSMUST00000237165.2 ENSMUST00000235892.2 ENSMUST00000114455.3 |
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr10_-_71180763 | 3.92 |
ENSMUST00000045887.9
|
Cisd1
|
CDGSH iron sulfur domain 1 |
| chr15_+_76227695 | 3.92 |
ENSMUST00000023210.8
ENSMUST00000231045.2 |
Cyc1
|
cytochrome c-1 |
| chr5_-_123320767 | 3.87 |
ENSMUST00000154713.8
ENSMUST00000031398.14 |
Hpd
|
4-hydroxyphenylpyruvic acid dioxygenase |
| chr9_-_107546166 | 3.81 |
ENSMUST00000177567.8
|
Slc38a3
|
solute carrier family 38, member 3 |
| chr13_-_74498320 | 3.78 |
ENSMUST00000221594.2
ENSMUST00000022062.8 |
Sdha
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr9_-_107546195 | 3.73 |
ENSMUST00000192990.6
|
Slc38a3
|
solute carrier family 38, member 3 |
| chr7_-_140856642 | 3.68 |
ENSMUST00000080654.7
ENSMUST00000167263.9 |
Cdhr5
|
cadherin-related family member 5 |
| chr7_+_26821266 | 3.52 |
ENSMUST00000206552.2
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2 |
| chr3_+_32791139 | 3.48 |
ENSMUST00000127477.8
ENSMUST00000121778.8 ENSMUST00000154257.8 |
Ndufb5
|
NADH:ubiquinone oxidoreductase subunit B5 |
| chr2_-_25391729 | 3.47 |
ENSMUST00000015227.4
|
C8g
|
complement component 8, gamma polypeptide |
| chr19_-_3962733 | 3.46 |
ENSMUST00000075092.8
ENSMUST00000235847.2 ENSMUST00000235301.2 ENSMUST00000237341.2 |
Ndufs8
|
NADH:ubiquinone oxidoreductase core subunit S8 |
| chr11_+_98932062 | 3.43 |
ENSMUST00000017637.13
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr8_-_13544478 | 3.35 |
ENSMUST00000033828.7
|
Gas6
|
growth arrest specific 6 |
| chr15_+_3300249 | 3.33 |
ENSMUST00000082424.12
ENSMUST00000159158.9 ENSMUST00000159216.10 ENSMUST00000160311.3 |
Selenop
|
selenoprotein P |
| chr5_-_147259245 | 3.31 |
ENSMUST00000100433.5
|
Urad
|
ureidoimidazoline (2-oxo-4-hydroxy-4-carboxy-5) decarboxylase |
| chr18_-_10706701 | 3.29 |
ENSMUST00000002549.9
ENSMUST00000117726.9 ENSMUST00000117828.9 |
Abhd3
|
abhydrolase domain containing 3 |
| chr9_-_106125055 | 3.22 |
ENSMUST00000074082.13
|
Alas1
|
aminolevulinic acid synthase 1 |
| chr5_+_114284585 | 3.22 |
ENSMUST00000102582.8
|
Acacb
|
acetyl-Coenzyme A carboxylase beta |
| chr7_-_81356653 | 3.09 |
ENSMUST00000026922.15
|
Homer2
|
homer scaffolding protein 2 |
| chr13_+_84370405 | 3.06 |
ENSMUST00000057495.10
ENSMUST00000225069.2 |
Tmem161b
|
transmembrane protein 161B |
| chr9_-_70048766 | 3.05 |
ENSMUST00000034749.16
|
Fam81a
|
family with sequence similarity 81, member A |
| chr9_+_107957621 | 3.05 |
ENSMUST00000035211.14
|
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr7_-_81356557 | 3.03 |
ENSMUST00000207983.2
|
Homer2
|
homer scaffolding protein 2 |
| chr1_-_175453117 | 3.02 |
ENSMUST00000027810.14
|
Fh1
|
fumarate hydratase 1 |
| chr11_-_53321606 | 3.00 |
ENSMUST00000061326.5
ENSMUST00000109021.4 |
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr9_+_107957640 | 3.00 |
ENSMUST00000162886.2
|
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr6_-_113508536 | 2.95 |
ENSMUST00000032425.7
|
Emc3
|
ER membrane protein complex subunit 3 |
| chr15_-_96947963 | 2.93 |
ENSMUST00000230907.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr8_+_95564949 | 2.90 |
ENSMUST00000034234.15
ENSMUST00000159871.4 |
Coq9
|
coenzyme Q9 |
| chr11_-_11848107 | 2.86 |
ENSMUST00000178704.8
|
Ddc
|
dopa decarboxylase |
| chr6_+_125297596 | 2.86 |
ENSMUST00000176655.8
ENSMUST00000176110.8 |
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr4_+_155646807 | 2.84 |
ENSMUST00000030939.14
|
Nadk
|
NAD kinase |
| chr11_-_11848044 | 2.82 |
ENSMUST00000066237.10
|
Ddc
|
dopa decarboxylase |
| chr2_-_157408239 | 2.82 |
ENSMUST00000109528.9
ENSMUST00000088494.3 |
Blcap
|
bladder cancer associated protein |
| chr2_+_103396638 | 2.80 |
ENSMUST00000076212.4
|
Abtb2
|
ankyrin repeat and BTB (POZ) domain containing 2 |
| chr7_-_48494959 | 2.79 |
ENSMUST00000208050.2
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr13_-_86194889 | 2.78 |
ENSMUST00000131011.2
|
Cox7c
|
cytochrome c oxidase subunit 7C |
| chr2_+_155359868 | 2.76 |
ENSMUST00000029135.15
ENSMUST00000065973.9 |
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr9_+_55234197 | 2.74 |
ENSMUST00000085754.10
ENSMUST00000034862.5 |
Tmem266
|
transmembrane protein 266 |
| chr17_-_56424265 | 2.74 |
ENSMUST00000113072.3
|
Plin5
|
perilipin 5 |
| chr3_-_90421557 | 2.72 |
ENSMUST00000107340.2
ENSMUST00000060738.9 |
S100a1
|
S100 calcium binding protein A1 |
| chr14_-_20844074 | 2.71 |
ENSMUST00000080440.14
ENSMUST00000100837.11 ENSMUST00000071816.7 |
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr6_+_145156860 | 2.66 |
ENSMUST00000111725.8
ENSMUST00000111726.10 ENSMUST00000039729.5 ENSMUST00000111723.8 ENSMUST00000111724.8 ENSMUST00000111721.2 ENSMUST00000111719.2 |
Etfrf1
|
electron transfer flavoprotein regulatory factor 1 |
| chr4_+_62278932 | 2.66 |
ENSMUST00000084526.12
|
Slc31a1
|
solute carrier family 31, member 1 |
| chr11_-_94568228 | 2.64 |
ENSMUST00000116349.9
|
Xylt2
|
xylosyltransferase II |
| chr7_-_30755007 | 2.64 |
ENSMUST00000206474.2
ENSMUST00000205807.2 ENSMUST00000039909.13 ENSMUST00000206305.2 ENSMUST00000205439.2 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr17_-_56424577 | 2.60 |
ENSMUST00000019808.12
|
Plin5
|
perilipin 5 |
| chr18_+_9212154 | 2.55 |
ENSMUST00000041080.7
|
Fzd8
|
frizzled class receptor 8 |
| chr11_-_118139331 | 2.53 |
ENSMUST00000017276.14
|
Cyth1
|
cytohesin 1 |
| chr10_+_79552421 | 2.48 |
ENSMUST00000099513.8
ENSMUST00000020581.3 |
Hcn2
|
hyperpolarization-activated, cyclic nucleotide-gated K+ 2 |
| chr11_+_120375439 | 2.43 |
ENSMUST00000043627.8
|
Mrpl12
|
mitochondrial ribosomal protein L12 |
| chrX_-_51254129 | 2.38 |
ENSMUST00000033450.3
|
Gpc4
|
glypican 4 |
| chr4_-_129121676 | 2.38 |
ENSMUST00000106051.8
|
C77080
|
expressed sequence C77080 |
| chr7_-_126944578 | 2.37 |
ENSMUST00000060783.7
|
Zfp768
|
zinc finger protein 768 |
| chr19_-_6957789 | 2.33 |
ENSMUST00000070878.9
ENSMUST00000177752.9 |
Fkbp2
|
FK506 binding protein 2 |
| chr17_+_44263890 | 2.33 |
ENSMUST00000177857.9
ENSMUST00000044792.6 |
Rcan2
|
regulator of calcineurin 2 |
| chr1_+_72863641 | 2.31 |
ENSMUST00000047328.11
|
Igfbp2
|
insulin-like growth factor binding protein 2 |
| chr17_-_26087696 | 2.30 |
ENSMUST00000236479.2
ENSMUST00000235806.2 ENSMUST00000026828.7 |
Mcrip2
|
MAPK regulated corepressor interacting protein 2 |
| chr4_+_128887017 | 2.28 |
ENSMUST00000030583.13
ENSMUST00000102604.11 |
Ak2
|
adenylate kinase 2 |
| chr16_-_37474772 | 2.27 |
ENSMUST00000023514.4
|
Ndufb4
|
NADH:ubiquinone oxidoreductase subunit B4 |
| chr8_+_121395047 | 2.27 |
ENSMUST00000181795.2
|
Cox4i1
|
cytochrome c oxidase subunit 4I1 |
| chrX_-_47602395 | 2.26 |
ENSMUST00000114945.9
ENSMUST00000037349.8 |
Aifm1
|
apoptosis-inducing factor, mitochondrion-associated 1 |
| chr9_-_121686601 | 2.26 |
ENSMUST00000213124.2
ENSMUST00000215300.2 ENSMUST00000213147.2 |
Higd1a
|
HIG1 domain family, member 1A |
| chr11_+_74540284 | 2.25 |
ENSMUST00000117818.2
ENSMUST00000092915.12 |
Cluh
|
clustered mitochondria (cluA/CLU1) homolog |
| chr3_+_90421742 | 2.23 |
ENSMUST00000048138.8
|
S100a13
|
S100 calcium binding protein A13 |
| chr14_-_20844034 | 2.22 |
ENSMUST00000226630.2
|
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr9_-_121686643 | 2.20 |
ENSMUST00000215007.2
ENSMUST00000216914.2 |
Higd1a
|
HIG1 domain family, member 1A |
| chr7_+_83281167 | 2.17 |
ENSMUST00000075418.15
|
Stard5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr9_+_65536892 | 2.14 |
ENSMUST00000169003.8
|
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr7_-_126944754 | 2.09 |
ENSMUST00000205266.2
|
Zfp768
|
zinc finger protein 768 |
| chr4_-_19708910 | 2.09 |
ENSMUST00000108246.9
|
Wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr3_-_131096792 | 2.08 |
ENSMUST00000200236.2
ENSMUST00000106337.7 |
Cyp2u1
|
cytochrome P450, family 2, subfamily u, polypeptide 1 |
| chr8_-_71315902 | 2.07 |
ENSMUST00000212611.2
|
Kcnn1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
| chr18_+_77861656 | 2.05 |
ENSMUST00000114748.2
|
Atp5a1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1 |
| chr16_+_22769822 | 2.04 |
ENSMUST00000023590.9
|
Hrg
|
histidine-rich glycoprotein |
| chr13_-_53083494 | 2.03 |
ENSMUST00000123599.8
|
Auh
|
AU RNA binding protein/enoyl-coenzyme A hydratase |
| chr13_+_51254852 | 1.98 |
ENSMUST00000095797.6
|
Spin1
|
spindlin 1 |
| chr13_-_67053384 | 1.98 |
ENSMUST00000021993.5
|
Uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chr19_+_4761181 | 1.97 |
ENSMUST00000008991.8
|
Sptbn2
|
spectrin beta, non-erythrocytic 2 |
| chr15_+_10224052 | 1.95 |
ENSMUST00000128450.8
ENSMUST00000148257.8 ENSMUST00000128921.8 |
Prlr
|
prolactin receptor |
| chr2_-_84508385 | 1.93 |
ENSMUST00000189772.2
ENSMUST00000053664.9 ENSMUST00000111664.8 |
Gm28635
Tmx2
|
predicted gene 28635 thioredoxin-related transmembrane protein 2 |
| chr11_+_115353290 | 1.93 |
ENSMUST00000106532.4
ENSMUST00000092445.12 ENSMUST00000153466.2 |
Slc16a5
|
solute carrier family 16 (monocarboxylic acid transporters), member 5 |
| chr9_+_107217786 | 1.92 |
ENSMUST00000042581.4
|
6430571L13Rik
|
RIKEN cDNA 6430571L13 gene |
| chr4_-_6275629 | 1.90 |
ENSMUST00000029905.2
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr2_-_73741664 | 1.90 |
ENSMUST00000111996.8
ENSMUST00000018914.3 |
Atp5g3
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) |
| chr1_+_4878046 | 1.89 |
ENSMUST00000027036.11
ENSMUST00000150971.8 ENSMUST00000119612.9 ENSMUST00000137887.8 ENSMUST00000115529.8 |
Lypla1
|
lysophospholipase 1 |
| chr1_+_74324089 | 1.88 |
ENSMUST00000113805.8
ENSMUST00000027370.13 ENSMUST00000087226.11 |
Pnkd
|
paroxysmal nonkinesiogenic dyskinesia |
| chr11_+_75084609 | 1.88 |
ENSMUST00000102514.4
|
Rtn4rl1
|
reticulon 4 receptor-like 1 |
| chr16_+_22769844 | 1.88 |
ENSMUST00000232422.2
|
Hrg
|
histidine-rich glycoprotein |
| chr10_-_78300802 | 1.86 |
ENSMUST00000041616.15
|
Pdxk
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr8_-_125296435 | 1.86 |
ENSMUST00000238882.2
ENSMUST00000063278.7 |
Agt
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8) |
| chr8_-_70975734 | 1.82 |
ENSMUST00000137610.3
|
Kxd1
|
KxDL motif containing 1 |
| chr9_-_21913833 | 1.80 |
ENSMUST00000115336.10
|
Odad3
|
outer dynein arm docking complex subunit 3 |
| chr16_+_10363203 | 1.78 |
ENSMUST00000115824.10
|
Clec16a
|
C-type lectin domain family 16, member A |
| chr1_-_175319842 | 1.78 |
ENSMUST00000195324.6
ENSMUST00000192227.6 ENSMUST00000194555.6 |
Rgs7
|
regulator of G protein signaling 7 |
| chr16_+_10363222 | 1.77 |
ENSMUST00000155633.8
ENSMUST00000066345.15 |
Clec16a
|
C-type lectin domain family 16, member A |
| chr9_-_44714263 | 1.77 |
ENSMUST00000044694.8
|
Ttc36
|
tetratricopeptide repeat domain 36 |
| chr9_+_65538352 | 1.75 |
ENSMUST00000216342.2
ENSMUST00000216382.2 |
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr11_-_118139312 | 1.74 |
ENSMUST00000100181.11
|
Cyth1
|
cytohesin 1 |
| chr11_-_118139418 | 1.72 |
ENSMUST00000106305.9
|
Cyth1
|
cytohesin 1 |
| chr6_+_71176811 | 1.71 |
ENSMUST00000067492.8
|
Fabp1
|
fatty acid binding protein 1, liver |
| chr5_-_87572060 | 1.71 |
ENSMUST00000072818.6
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr9_-_50515089 | 1.70 |
ENSMUST00000000175.6
|
Sdhd
|
succinate dehydrogenase complex, subunit D, integral membrane protein |
| chr18_+_77273510 | 1.70 |
ENSMUST00000075290.8
ENSMUST00000079618.11 |
St8sia5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr14_+_79086665 | 1.69 |
ENSMUST00000227255.2
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr4_-_148244028 | 1.69 |
ENSMUST00000167160.8
ENSMUST00000151246.8 |
Fbxo44
|
F-box protein 44 |
| chr7_+_119217004 | 1.69 |
ENSMUST00000047929.13
ENSMUST00000135683.3 |
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr16_+_10363254 | 1.65 |
ENSMUST00000115827.8
ENSMUST00000150894.2 ENSMUST00000038145.13 |
Clec16a
|
C-type lectin domain family 16, member A |
| chr3_+_96484294 | 1.65 |
ENSMUST00000148290.2
|
Gm16253
|
predicted gene 16253 |
| chr9_-_21913896 | 1.65 |
ENSMUST00000044926.6
|
Odad3
|
outer dynein arm docking complex subunit 3 |
| chr11_+_75400889 | 1.65 |
ENSMUST00000042972.7
|
Rilp
|
Rab interacting lysosomal protein |
| chr9_-_63306497 | 1.64 |
ENSMUST00000168665.3
|
2300009A05Rik
|
RIKEN cDNA 2300009A05 gene |
| chrX_-_71699740 | 1.61 |
ENSMUST00000055966.13
|
Gabra3
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 3 |
| chr4_-_8239034 | 1.61 |
ENSMUST00000066674.8
|
Car8
|
carbonic anhydrase 8 |
| chr19_-_34856853 | 1.61 |
ENSMUST00000036584.13
|
Pank1
|
pantothenate kinase 1 |
| chr4_+_138161958 | 1.61 |
ENSMUST00000044058.11
ENSMUST00000105813.8 ENSMUST00000105815.2 |
Mul1
|
mitochondrial ubiquitin ligase activator of NFKB 1 |
| chr15_+_68800261 | 1.60 |
ENSMUST00000022954.7
|
Khdrbs3
|
KH domain containing, RNA binding, signal transduction associated 3 |
| chr16_+_22926162 | 1.60 |
ENSMUST00000023599.13
ENSMUST00000168891.8 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr16_+_22926504 | 1.60 |
ENSMUST00000187168.7
ENSMUST00000232287.2 ENSMUST00000077605.12 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr15_-_76191301 | 1.59 |
ENSMUST00000171340.9
ENSMUST00000023222.13 ENSMUST00000164189.2 |
Oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr7_-_4525793 | 1.59 |
ENSMUST00000140424.8
|
Tnni3
|
troponin I, cardiac 3 |
| chr11_-_116089595 | 1.59 |
ENSMUST00000072948.11
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr4_-_122854966 | 1.59 |
ENSMUST00000030408.12
ENSMUST00000127047.2 |
Mfsd2a
|
major facilitator superfamily domain containing 2A |
| chr6_+_91134358 | 1.57 |
ENSMUST00000155007.2
|
Hdac11
|
histone deacetylase 11 |
| chr4_+_140970161 | 1.56 |
ENSMUST00000138096.8
ENSMUST00000006618.9 ENSMUST00000125392.8 |
Arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr19_-_47079052 | 1.56 |
ENSMUST00000235771.2
ENSMUST00000096014.5 ENSMUST00000236170.2 |
Atp5md
|
ATP synthase membrane subunit DAPIT |
| chr11_+_98239230 | 1.56 |
ENSMUST00000078694.13
|
Ppp1r1b
|
protein phosphatase 1, regulatory inhibitor subunit 1B |
| chr16_-_23339548 | 1.54 |
ENSMUST00000089883.7
|
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr2_-_102903680 | 1.54 |
ENSMUST00000132449.8
ENSMUST00000111183.2 ENSMUST00000011058.9 |
Pdhx
|
pyruvate dehydrogenase complex, component X |
| chr7_-_80053063 | 1.53 |
ENSMUST00000147150.2
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr6_+_91133647 | 1.53 |
ENSMUST00000041736.11
|
Hdac11
|
histone deacetylase 11 |
| chr11_-_72157419 | 1.51 |
ENSMUST00000208056.2
ENSMUST00000208912.2 ENSMUST00000140167.3 ENSMUST00000137701.3 |
Slc13a5
|
solute carrier family 13 (sodium-dependent citrate transporter), member 5 |
| chr11_-_116089866 | 1.51 |
ENSMUST00000066587.12
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr11_-_96720309 | 1.51 |
ENSMUST00000167149.8
|
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1 |
| chr4_-_49408040 | 1.50 |
ENSMUST00000081541.9
|
Acnat2
|
acyl-coenzyme A amino acid N-acyltransferase 2 |
| chr14_+_79086492 | 1.48 |
ENSMUST00000040990.7
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr8_+_121394961 | 1.47 |
ENSMUST00000034276.13
ENSMUST00000181586.8 |
Cox4i1
|
cytochrome c oxidase subunit 4I1 |
| chr6_+_91133755 | 1.47 |
ENSMUST00000143621.8
|
Hdac11
|
histone deacetylase 11 |
| chr19_+_3372296 | 1.44 |
ENSMUST00000237938.2
|
Cpt1a
|
carnitine palmitoyltransferase 1a, liver |
| chr4_-_83242490 | 1.41 |
ENSMUST00000102823.10
|
Ttc39b
|
tetratricopeptide repeat domain 39B |
| chr2_+_166647426 | 1.41 |
ENSMUST00000099078.10
|
Arfgef2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) |
| chr12_-_71183371 | 1.39 |
ENSMUST00000221367.2
ENSMUST00000220482.2 ENSMUST00000221892.2 ENSMUST00000221178.2 ENSMUST00000221559.2 ENSMUST00000166120.9 ENSMUST00000021486.10 ENSMUST00000221797.2 ENSMUST00000221815.2 |
Timm9
|
translocase of inner mitochondrial membrane 9 |
| chr9_-_106562852 | 1.38 |
ENSMUST00000169068.8
|
Tex264
|
testis expressed gene 264 |
| chr15_-_83989801 | 1.37 |
ENSMUST00000229826.2
ENSMUST00000082365.6 |
Sult4a1
|
sulfotransferase family 4A, member 1 |
| chr7_+_127399776 | 1.36 |
ENSMUST00000046863.12
ENSMUST00000206674.2 ENSMUST00000106272.8 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr9_-_106124917 | 1.35 |
ENSMUST00000112524.9
ENSMUST00000219129.2 |
Alas1
|
aminolevulinic acid synthase 1 |
| chr7_+_29883611 | 1.33 |
ENSMUST00000208441.2
|
Cox7a1
|
cytochrome c oxidase subunit 7A1 |
| chr5_-_23881353 | 1.32 |
ENSMUST00000198661.5
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr11_-_60702081 | 1.32 |
ENSMUST00000018744.15
|
Shmt1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr19_-_7194912 | 1.32 |
ENSMUST00000039758.6
|
Cox8a
|
cytochrome c oxidase subunit 8A |
| chr15_+_100659729 | 1.29 |
ENSMUST00000161564.2
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8 |
| chr15_-_77813123 | 1.29 |
ENSMUST00000109748.9
ENSMUST00000109747.9 ENSMUST00000100486.6 ENSMUST00000005487.12 |
Txn2
|
thioredoxin 2 |
| chr11_-_59118988 | 1.29 |
ENSMUST00000163300.8
ENSMUST00000061242.8 |
Arf1
|
ADP-ribosylation factor 1 |
| chr2_-_10084866 | 1.29 |
ENSMUST00000130067.2
ENSMUST00000139810.8 |
Atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr6_+_39569502 | 1.28 |
ENSMUST00000135671.8
ENSMUST00000119379.2 |
Ndufb2
|
NADH:ubiquinone oxidoreductase subunit B2 |
| chr11_+_120382666 | 1.28 |
ENSMUST00000026899.4
|
Slc25a10
|
solute carrier family 25 (mitochondrial carrier, dicarboxylate transporter), member 10 |
| chr10_+_127919142 | 1.27 |
ENSMUST00000026459.6
|
Atp5b
|
ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit |
| chr5_-_25200745 | 1.27 |
ENSMUST00000076306.12
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr2_-_160169414 | 1.26 |
ENSMUST00000099127.3
|
Gm826
|
predicted gene 826 |
| chr7_+_127399789 | 1.26 |
ENSMUST00000125188.8
|
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr1_-_134163102 | 1.26 |
ENSMUST00000187631.2
ENSMUST00000038191.8 ENSMUST00000086465.6 |
Adora1
|
adenosine A1 receptor |
| chr13_-_25121568 | 1.25 |
ENSMUST00000037615.7
|
Aldh5a1
|
aldhehyde dehydrogenase family 5, subfamily A1 |
| chr2_+_140012560 | 1.24 |
ENSMUST00000044825.5
|
Ndufaf5
|
NADH:ubiquinone oxidoreductase complex assembly factor 5 |
| chr5_+_150042092 | 1.24 |
ENSMUST00000200960.4
ENSMUST00000202530.4 |
Fry
|
FRY microtubule binding protein |
| chr1_+_153300874 | 1.24 |
ENSMUST00000042373.12
|
Shcbp1l
|
Shc SH2-domain binding protein 1-like |
| chr8_-_110305672 | 1.24 |
ENSMUST00000074898.8
|
Hp
|
haptoglobin |
| chr15_+_74435587 | 1.23 |
ENSMUST00000185682.7
ENSMUST00000170845.8 ENSMUST00000187599.2 |
Adgrb1
|
adhesion G protein-coupled receptor B1 |
| chr11_-_72157450 | 1.22 |
ENSMUST00000021161.14
|
Slc13a5
|
solute carrier family 13 (sodium-dependent citrate transporter), member 5 |
| chr10_-_42354276 | 1.21 |
ENSMUST00000151747.8
|
Afg1l
|
AFG1 like ATPase |
| chr4_-_40279382 | 1.21 |
ENSMUST00000108108.3
ENSMUST00000095128.10 |
Ndufb6
|
NADH:ubiquinone oxidoreductase subunit B6 |
| chr7_-_96981221 | 1.21 |
ENSMUST00000139582.9
|
Usp35
|
ubiquitin specific peptidase 35 |
| chr7_-_119122681 | 1.20 |
ENSMUST00000033267.4
|
Pdilt
|
protein disulfide isomerase-like, testis expressed |
| chr7_-_16761790 | 1.19 |
ENSMUST00000003183.12
|
Ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr9_-_110709175 | 1.18 |
ENSMUST00000050958.9
|
Tmie
|
transmembrane inner ear |
| chr4_+_150321659 | 1.18 |
ENSMUST00000133839.8
|
Eno1
|
enolase 1, alpha non-neuron |
| chr5_+_30437579 | 1.17 |
ENSMUST00000145167.9
|
Selenoi
|
selenoprotein I |
| chr11_+_70735572 | 1.17 |
ENSMUST00000076270.13
ENSMUST00000179114.8 ENSMUST00000100928.11 |
Rabep1
|
rabaptin, RAB GTPase binding effector protein 1 |
| chr4_+_150321272 | 1.17 |
ENSMUST00000080926.13
|
Eno1
|
enolase 1, alpha non-neuron |
| chr18_-_79152504 | 1.16 |
ENSMUST00000025430.11
|
Setbp1
|
SET binding protein 1 |
| chr4_-_131871797 | 1.16 |
ENSMUST00000056336.2
|
Oprd1
|
opioid receptor, delta 1 |
| chr9_-_21996693 | 1.16 |
ENSMUST00000179422.8
ENSMUST00000098937.10 ENSMUST00000177967.2 ENSMUST00000180180.8 |
Ecsit
|
ECSIT signalling integrator |
Network of associatons between targets according to the STRING database.
First level regulatory network of Esrrb_Esrra
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 21.3 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 2.5 | 7.5 | GO:2000487 | asparagine transport(GO:0006867) positive regulation of glutamine transport(GO:2000487) |
| 2.5 | 9.9 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 2.2 | 6.7 | GO:0009397 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 2.1 | 12.4 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 1.7 | 5.2 | GO:1904766 | negative regulation of macroautophagy by TORC1 signaling(GO:1904766) |
| 1.4 | 9.5 | GO:0015886 | heme transport(GO:0015886) |
| 1.3 | 9.4 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 1.3 | 3.8 | GO:2001293 | fatty-acyl-CoA biosynthetic process(GO:0046949) malonyl-CoA metabolic process(GO:2001293) |
| 1.2 | 3.5 | GO:0090420 | naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 1.1 | 5.5 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 1.1 | 6.5 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 1.1 | 5.3 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 1.0 | 4.0 | GO:0015744 | succinate transport(GO:0015744) |
| 0.9 | 2.8 | GO:1903920 | positive regulation of actin filament severing(GO:1903920) |
| 0.9 | 2.8 | GO:0019542 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 0.8 | 3.3 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.8 | 14.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.8 | 5.7 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.8 | 4.0 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.7 | 10.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.7 | 3.4 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.7 | 6.0 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.7 | 1.3 | GO:0021941 | negative regulation of cerebellar granule cell precursor proliferation(GO:0021941) |
| 0.7 | 2.6 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.7 | 4.0 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.6 | 3.7 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.6 | 1.9 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.6 | 1.9 | GO:0061461 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine import into cell(GO:1903410) |
| 0.6 | 3.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.6 | 4.2 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.6 | 1.8 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.6 | 6.4 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.6 | 5.7 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.5 | 3.3 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.5 | 1.6 | GO:0051977 | lysophospholipid transport(GO:0051977) |
| 0.5 | 3.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.5 | 4.4 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.5 | 1.8 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.4 | 1.3 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.4 | 1.3 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.4 | 0.9 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.4 | 3.9 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.4 | 1.3 | GO:0070256 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.4 | 1.3 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.4 | 3.3 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.4 | 1.2 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.4 | 1.2 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.4 | 1.2 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.4 | 1.5 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.4 | 1.9 | GO:0070859 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.4 | 2.7 | GO:0072719 | cellular response to cisplatin(GO:0072719) copper ion import across plasma membrane(GO:0098705) copper ion import into cell(GO:1902861) |
| 0.4 | 2.3 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.4 | 17.5 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.4 | 0.7 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.4 | 2.5 | GO:0071321 | cellular response to cGMP(GO:0071321) potassium ion import across plasma membrane(GO:1990573) |
| 0.3 | 2.0 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.3 | 3.6 | GO:1903278 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.3 | 9.0 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.3 | 3.5 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.3 | 1.0 | GO:0046108 | uridine catabolic process(GO:0006218) uridine metabolic process(GO:0046108) |
| 0.3 | 2.8 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.3 | 1.6 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.3 | 1.2 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.3 | 1.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.3 | 2.1 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.3 | 1.4 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.3 | 1.4 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.3 | 0.8 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.3 | 1.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.3 | 1.6 | GO:1901526 | negative regulation of defense response to virus by host(GO:0050689) regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.3 | 2.6 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.3 | 1.0 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.3 | 6.1 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.3 | 0.8 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.2 | 2.2 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.2 | 0.7 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.2 | 0.7 | GO:0060450 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) positive regulation of hindgut contraction(GO:0060450) |
| 0.2 | 3.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.2 | 1.9 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.2 | 1.0 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.2 | 2.4 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.2 | 3.6 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.2 | 0.9 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 | 3.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 0.9 | GO:0050823 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.2 | 0.7 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.2 | 0.7 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.2 | 3.0 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.2 | 0.9 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.2 | 0.9 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.2 | 5.5 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.2 | 1.0 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.2 | 0.8 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.2 | 7.2 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 2.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.2 | 2.9 | GO:0035376 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.2 | 0.6 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.2 | 2.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 0.4 | GO:0032730 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.2 | 1.8 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 0.5 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.2 | 0.7 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.2 | 17.3 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.2 | 1.4 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.2 | 2.5 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.2 | 3.0 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.2 | 1.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.2 | 0.7 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 0.2 | 2.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 0.8 | GO:0072363 | regulation of glycolytic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072363) |
| 0.2 | 0.5 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.2 | 0.9 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.2 | 0.8 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.2 | 0.6 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.2 | 0.5 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.1 | 0.3 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.1 | 0.7 | GO:0045964 | negative regulation of serotonin secretion(GO:0014063) positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.1 | 0.4 | GO:0046166 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 1.8 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.4 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.1 | 0.9 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 1.8 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.9 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 0.5 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 3.2 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.1 | 1.9 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.1 | 0.4 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 0.6 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.4 | GO:0009188 | ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.1 | 4.1 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.1 | 1.6 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.4 | GO:0046337 | phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.1 | 1.0 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.7 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 2.0 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 2.7 | GO:0090201 | negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.1 | 0.7 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 2.7 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.1 | 0.5 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.1 | 3.0 | GO:0007614 | short-term memory(GO:0007614) |
| 0.1 | 0.6 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.1 | 0.7 | GO:0015824 | proline transport(GO:0015824) |
| 0.1 | 1.8 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 2.0 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 3.3 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.1 | 0.4 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.1 | 1.9 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 0.5 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.1 | 2.0 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 0.3 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.1 | 1.7 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.3 | GO:0021852 | pyramidal neuron migration(GO:0021852) |
| 0.1 | 0.9 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 1.1 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.1 | 2.8 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.1 | 1.0 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.1 | 2.2 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) response to increased oxygen levels(GO:0036296) |
| 0.1 | 0.9 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.5 | GO:0060336 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.1 | 2.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.5 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.1 | 0.3 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.1 | 4.6 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.1 | 0.6 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.1 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) |
| 0.1 | 4.5 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.4 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.9 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 0.7 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.1 | 0.4 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.1 | 2.8 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.1 | 0.6 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 0.5 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.4 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.1 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.1 | 1.0 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 0.6 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 0.3 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 0.6 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.3 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 0.2 | GO:0033577 | protein glycosylation in endoplasmic reticulum(GO:0033577) |
| 0.1 | 0.3 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.1 | 0.3 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.1 | 1.4 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 1.0 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.1 | 0.2 | GO:0060112 | positive regulation of luteinizing hormone secretion(GO:0033686) generation of ovulation cycle rhythm(GO:0060112) |
| 0.1 | 0.5 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 2.9 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.9 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) cristae formation(GO:0042407) |
| 0.1 | 1.3 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.1 | 0.3 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 0.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.9 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 0.3 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 2.0 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 0.3 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 0.1 | 0.1 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.1 | 0.4 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.5 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.1 | 0.2 | GO:0033239 | negative regulation of cellular amine metabolic process(GO:0033239) negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.1 | 0.2 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.1 | 0.7 | GO:0070255 | regulation of mucus secretion(GO:0070255) |
| 0.1 | 2.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.8 | GO:0061525 | hindgut development(GO:0061525) |
| 0.1 | 0.3 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 2.1 | GO:0006110 | regulation of glycolytic process(GO:0006110) |
| 0.1 | 1.3 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.1 | 0.6 | GO:0045348 | positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 0.1 | 0.2 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.1 | 0.6 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 0.9 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 1.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 1.0 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.5 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) |
| 0.1 | 0.6 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.0 | 0.7 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.0 | 0.5 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.5 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.4 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 1.3 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.0 | 0.1 | GO:1904349 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) positive regulation of eating behavior(GO:1904000) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) regulation of small intestine smooth muscle contraction(GO:1904347) positive regulation of small intestine smooth muscle contraction(GO:1904349) gastric mucosal blood circulation(GO:1990768) small intestine smooth muscle contraction(GO:1990770) negative regulation of energy homeostasis(GO:2000506) |
| 0.0 | 0.9 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.8 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.7 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.2 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.0 | 1.1 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 1.3 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.4 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.0 | 0.9 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 1.8 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.7 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 2.6 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.6 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 | 0.8 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.2 | GO:1902163 | negative regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902163) |
| 0.0 | 1.4 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.5 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.0 | 0.9 | GO:0043153 | photoperiodism(GO:0009648) entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 2.2 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 2.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.5 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.4 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.5 | GO:0044065 | regulation of respiratory system process(GO:0044065) |
| 0.0 | 1.0 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 2.5 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 1.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 2.2 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.1 | GO:0072014 | proximal tubule development(GO:0072014) regulation of androgen receptor activity(GO:2000823) |
| 0.0 | 0.3 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.3 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.6 | GO:0032674 | regulation of interleukin-5 production(GO:0032674) |
| 0.0 | 0.9 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.6 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.4 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.0 | 0.5 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.3 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.2 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.1 | GO:0036394 | amylase secretion(GO:0036394) |
| 0.0 | 0.6 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 0.3 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 1.9 | GO:0043507 | positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.4 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.0 | 1.8 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.8 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 1.1 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.3 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 | 0.2 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 1.2 | GO:0030816 | positive regulation of cAMP metabolic process(GO:0030816) |
| 0.0 | 1.0 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.7 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.7 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.4 | GO:0045823 | positive regulation of heart contraction(GO:0045823) |
| 0.0 | 0.3 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.3 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.2 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.0 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.2 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.2 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.0 | 0.6 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) |
| 0.0 | 0.3 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.6 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.2 | GO:0002029 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) |
| 0.0 | 0.1 | GO:0071231 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.0 | 0.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.7 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.8 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 0.2 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.3 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.3 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 0.1 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.1 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.6 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.1 | GO:0002827 | positive regulation of T-helper 1 type immune response(GO:0002827) |
| 0.0 | 1.1 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.0 | 0.2 | GO:1903543 | regulation of exosomal secretion(GO:1903541) positive regulation of exosomal secretion(GO:1903543) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.5 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 1.7 | 5.2 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 1.3 | 3.9 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.9 | 10.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.9 | 9.9 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.8 | 2.5 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.6 | 2.5 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.6 | 9.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.6 | 5.1 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.5 | 2.7 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.4 | 1.2 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.4 | 3.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.4 | 2.3 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.4 | 2.6 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.4 | 3.7 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.4 | 1.4 | GO:0032280 | axonemal microtubule(GO:0005879) symmetric synapse(GO:0032280) |
| 0.3 | 5.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.3 | 4.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.3 | 22.4 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.3 | 1.9 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.2 | 1.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 2.0 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 3.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 1.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.2 | 2.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.2 | 0.7 | GO:0005757 | mitochondrial permeability transition pore complex(GO:0005757) |
| 0.2 | 0.5 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 0.5 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.2 | 0.9 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.2 | 1.9 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.2 | 0.7 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.2 | 1.9 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 2.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 0.7 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.2 | 4.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.2 | 2.6 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.2 | 8.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 3.8 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 6.8 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.4 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 0.6 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.1 | 0.7 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 0.8 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.9 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) TAP complex(GO:0042825) |
| 0.1 | 0.3 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 4.0 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.1 | 0.9 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 1.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.9 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 11.9 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.1 | 0.3 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.1 | 1.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.0 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.4 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.1 | 1.5 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 1.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.4 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.1 | 5.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 22.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 1.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 1.4 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 18.6 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 6.2 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.1 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.1 | 0.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.6 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 0.5 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.1 | 1.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 5.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 6.6 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 15.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 3.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 2.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.1 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 3.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.4 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 1.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 1.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.3 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.7 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 0.3 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 2.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.7 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.5 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 4.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 5.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 2.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 5.2 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 2.6 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.4 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 2.2 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 17.7 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 18.3 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 4.5 | 22.3 | GO:0004021 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 4.2 | 16.9 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 3.2 | 9.5 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 2.5 | 9.9 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 2.2 | 6.7 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 2.1 | 12.4 | GO:0004359 | glutaminase activity(GO:0004359) |
| 1.9 | 7.5 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 1.6 | 9.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.5 | 4.6 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 1.4 | 5.7 | GO:0036468 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 1.3 | 3.9 | GO:0008127 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) quercetin 2,3-dioxygenase activity(GO:0008127) |
| 1.0 | 4.0 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.9 | 8.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.9 | 5.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.9 | 2.6 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.9 | 6.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.8 | 4.2 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.8 | 3.3 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.8 | 4.7 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.8 | 3.8 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.7 | 4.0 | GO:0016019 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.7 | 2.6 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 0.6 | 5.2 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.6 | 1.9 | GO:0070279 | vitamin B6 binding(GO:0070279) |
| 0.6 | 1.7 | GO:0016213 | linoleoyl-CoA desaturase activity(GO:0016213) |
| 0.6 | 5.7 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.6 | 3.9 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.6 | 2.8 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.5 | 2.6 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.5 | 1.6 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.5 | 1.4 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.5 | 0.9 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.5 | 1.8 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.5 | 13.3 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.4 | 1.3 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 0.4 | 6.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.4 | 4.6 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.4 | 2.5 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.4 | 5.3 | GO:0035473 | lipase binding(GO:0035473) |
| 0.4 | 2.8 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.4 | 2.0 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.4 | 1.2 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 0.4 | 1.9 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.4 | 2.6 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.4 | 6.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.3 | 1.0 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.3 | 1.3 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.3 | 3.6 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.3 | 2.4 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.3 | 1.2 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.3 | 1.8 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.3 | 1.6 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.3 | 2.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 3.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 3.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 2.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 2.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.2 | 1.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 1.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.2 | 1.7 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.2 | 2.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 0.8 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.2 | 6.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 1.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.2 | 3.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.2 | 1.9 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.2 | 0.4 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.2 | 0.6 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.2 | 0.7 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.2 | 2.7 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.2 | 0.5 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.2 | 1.6 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.2 | 1.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.2 | 1.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 0.7 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.2 | 1.9 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 0.5 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.2 | 3.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 3.9 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.2 | 2.0 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.2 | 7.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 3.5 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.2 | 0.6 | GO:0004104 | choline kinase activity(GO:0004103) cholinesterase activity(GO:0004104) |
| 0.2 | 1.0 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.2 | 1.0 | GO:0070004 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.2 | 0.8 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.2 | 0.9 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.2 | 3.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.9 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.1 | 1.0 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 2.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 1.3 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 4.0 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.4 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.1 | 0.4 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.1 | 0.4 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 2.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.9 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.1 | 2.6 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.4 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.1 | 0.7 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 1.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.6 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.1 | 1.8 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.1 | 0.8 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 2.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.7 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 2.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.3 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.1 | 0.9 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.3 | GO:0099530 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 1.6 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.1 | 0.3 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.1 | 0.5 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 0.6 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 2.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.5 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 0.1 | 2.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.3 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 0.3 | GO:0004615 | phosphomannomutase activity(GO:0004615) |
| 0.1 | 1.9 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 2.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.6 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.1 | 0.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 19.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.1 | GO:0008556 | potassium-transporting ATPase activity(GO:0008556) |
| 0.1 | 0.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.5 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 0.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 1.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 2.1 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 2.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.7 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 3.0 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.5 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 2.1 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 0.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.4 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 1.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 1.9 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.1 | 0.7 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 0.2 | GO:0004133 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.1 | 0.3 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.1 | 2.2 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) |
| 0.1 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.8 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 2.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.9 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 3.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.7 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.1 | 0.6 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 1.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.8 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.3 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.3 | GO:0005186 | pheromone activity(GO:0005186) |
| 0.1 | 0.4 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 2.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 0.8 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 3.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 1.5 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 2.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.4 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 1.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.3 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.0 | 1.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 1.0 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.2 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.0 | 0.7 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.9 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 1.0 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.7 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 5.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 1.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 5.6 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 1.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 2.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 4.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 2.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 1.0 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 1.7 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.5 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 1.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.9 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 1.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.0 | 1.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.9 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.3 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 1.2 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.4 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 0.0 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.9 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 1.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 15.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 6.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 6.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.3 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 6.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 0.9 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 4.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 4.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 1.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 8.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 10.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.7 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 2.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.2 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 3.8 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 0.8 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 53.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.9 | 19.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.8 | 12.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.6 | 46.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.5 | 9.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.4 | 7.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.3 | 4.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.3 | 5.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 2.8 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 4.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 7.0 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.2 | 21.2 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.2 | 3.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 4.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 2.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 2.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 1.8 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 2.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 3.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 3.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 2.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 2.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 3.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 14.7 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 0.9 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 3.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 4.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 0.8 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 3.1 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 0.4 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.1 | 1.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 4.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 0.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 2.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 2.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 0.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.3 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.7 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 3.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.4 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 5.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.6 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 1.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.5 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 2.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 1.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 5.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.0 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 1.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.6 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 1.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.4 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.4 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 2.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.5 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 2.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.1 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |