Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
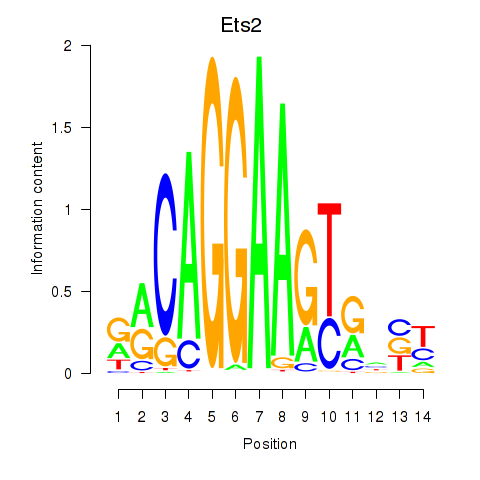
Results for Ets2
Z-value: 1.81
Transcription factors associated with Ets2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ets2
|
ENSMUSG00000022895.17 | E26 avian leukemia oncogene 2, 3' domain |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ets2 | mm39_v1_chr16_+_95502911_95502960 | -0.45 | 5.9e-03 | Click! |
Activity profile of Ets2 motif
Sorted Z-values of Ets2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Ets2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.7 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 4.3 | 29.8 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 4.0 | 4.0 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 3.9 | 11.8 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 2.6 | 46.6 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 2.3 | 7.0 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 2.2 | 13.3 | GO:0034627 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 2.0 | 6.0 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 1.9 | 5.8 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 1.8 | 5.5 | GO:0019085 | early viral transcription(GO:0019085) |
| 1.7 | 10.4 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 1.7 | 8.3 | GO:0045994 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 1.4 | 10.0 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 1.3 | 5.1 | GO:0007113 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 1.2 | 3.6 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 1.2 | 2.4 | GO:0019086 | late viral transcription(GO:0019086) |
| 1.2 | 7.1 | GO:1990839 | response to endothelin(GO:1990839) |
| 1.2 | 3.5 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 1.1 | 3.4 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 1.1 | 4.4 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 1.1 | 3.2 | GO:0046038 | GMP catabolic process(GO:0046038) |
| 1.1 | 3.2 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 1.1 | 3.2 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.9 | 1.8 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.9 | 3.6 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.9 | 3.5 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.8 | 4.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.8 | 7.0 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.8 | 3.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.8 | 3.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.8 | 4.6 | GO:0070425 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.8 | 3.0 | GO:0000105 | histidine biosynthetic process(GO:0000105) 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.7 | 16.3 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.7 | 1.4 | GO:0060737 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.7 | 6.4 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.7 | 2.1 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.7 | 2.6 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.6 | 5.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.6 | 3.4 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.6 | 1.7 | GO:1904582 | positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.6 | 5.7 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.5 | 1.1 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.5 | 7.0 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.5 | 15.5 | GO:0043574 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.5 | 5.2 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.5 | 1.6 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.5 | 2.6 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.5 | 2.6 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.5 | 1.5 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.5 | 1.5 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.5 | 2.0 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.5 | 1.5 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.5 | 4.4 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.5 | 5.1 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.5 | 2.7 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.4 | 1.7 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.4 | 9.9 | GO:0019363 | pyridine nucleotide biosynthetic process(GO:0019363) |
| 0.4 | 8.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.4 | 2.0 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.4 | 1.2 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.4 | 2.8 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.4 | 2.8 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.4 | 2.7 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.4 | 2.7 | GO:0072092 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.4 | 1.9 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.4 | 5.7 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.4 | 0.7 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.4 | 6.7 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.4 | 2.2 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.3 | 3.5 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.3 | 1.0 | GO:2000556 | regulation of T-helper 1 cell cytokine production(GO:2000554) positive regulation of T-helper 1 cell cytokine production(GO:2000556) |
| 0.3 | 5.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.3 | 3.1 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.3 | 1.0 | GO:0060729 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) intestinal epithelial structure maintenance(GO:0060729) |
| 0.3 | 1.7 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.3 | 3.0 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.3 | 3.3 | GO:2000535 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.3 | 1.3 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.3 | 0.7 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.3 | 3.5 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.3 | 5.6 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.3 | 1.8 | GO:0015867 | ATP transport(GO:0015867) |
| 0.3 | 1.8 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.3 | 5.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.3 | 1.2 | GO:0003142 | cardiogenic plate morphogenesis(GO:0003142) regulation of transcription from RNA polymerase II promoter involved in definitive endodermal cell fate specification(GO:0060807) |
| 0.3 | 1.2 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.3 | 3.4 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.3 | 2.5 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.3 | 1.4 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.3 | 3.3 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.3 | 1.1 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.3 | 9.8 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.3 | 2.1 | GO:0072642 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.3 | 1.6 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.3 | 4.7 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.3 | 3.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.3 | 1.3 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.3 | 1.5 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.3 | 0.8 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.2 | 6.8 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.2 | 2.4 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.2 | 5.9 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.2 | 2.8 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.2 | 2.6 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 0.9 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.2 | 0.9 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.2 | 1.5 | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) |
| 0.2 | 1.1 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) |
| 0.2 | 4.6 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.2 | 2.1 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.2 | 1.0 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.2 | 0.4 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.2 | 2.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 0.6 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.2 | 2.6 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.2 | 0.6 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.2 | 8.5 | GO:0015893 | drug transport(GO:0015893) |
| 0.2 | 0.8 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.2 | 1.2 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.2 | 28.3 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.2 | 2.6 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.2 | 1.3 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.2 | 2.2 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.2 | 3.7 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.2 | 1.2 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.2 | 1.0 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.2 | 1.5 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.2 | 2.2 | GO:0030238 | male sex determination(GO:0030238) |
| 0.2 | 5.1 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.2 | 2.6 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.2 | 3.8 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.2 | 2.4 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.2 | 0.6 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.2 | 2.8 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.2 | 1.8 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.2 | 2.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 0.6 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 1.3 | GO:0002349 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 | 2.1 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.1 | 6.2 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 2.1 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.1 | 3.5 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 1.0 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.3 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.1 | 3.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 1.5 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.1 | 1.7 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.1 | 0.4 | GO:1903027 | asymmetric Golgi ribbon formation(GO:0090164) regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.1 | 1.9 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 0.9 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 1.8 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 3.7 | GO:0051654 | establishment of mitochondrion localization(GO:0051654) |
| 0.1 | 6.7 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 1.2 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 1.0 | GO:1905206 | positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.1 | 1.3 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.1 | 1.6 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.8 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 0.9 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 0.9 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.1 | 0.6 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.1 | 0.8 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.1 | 1.3 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 0.5 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 4.2 | GO:0030811 | regulation of glycolytic process(GO:0006110) regulation of nucleotide catabolic process(GO:0030811) |
| 0.1 | 10.0 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 0.9 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.9 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 1.6 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 | 3.1 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 | 2.6 | GO:1903441 | protein localization to ciliary membrane(GO:1903441) |
| 0.1 | 0.6 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.1 | 0.5 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.1 | 8.2 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.1 | 2.4 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 0.7 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.1 | 2.2 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.1 | 3.6 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 0.9 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.1 | 1.5 | GO:0035743 | CD4-positive, alpha-beta T cell cytokine production(GO:0035743) T-helper 2 cell cytokine production(GO:0035745) |
| 0.1 | 1.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.4 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 2.6 | GO:0090662 | ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.1 | 2.0 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.3 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 1.2 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 0.6 | GO:1990009 | retinal cell apoptotic process(GO:1990009) |
| 0.1 | 2.2 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.1 | 2.5 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 1.0 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 1.2 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 1.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.1 | 1.6 | GO:0071236 | cellular response to antibiotic(GO:0071236) |
| 0.1 | 3.4 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 2.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 1.8 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.4 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 6.0 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 0.5 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.1 | 0.4 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.5 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 2.9 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 0.5 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 0.3 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 2.0 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 0.9 | GO:0003283 | atrial septum development(GO:0003283) |
| 0.1 | 0.1 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.7 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 0.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.4 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.1 | 1.6 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.1 | 3.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 1.2 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 1.0 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.6 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 2.6 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 4.1 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.1 | 1.5 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 | 2.0 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.7 | GO:0002827 | positive regulation of T-helper 1 type immune response(GO:0002827) |
| 0.1 | 1.0 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.1 | 0.2 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.6 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 1.0 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 4.8 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 1.6 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.4 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.8 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.2 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 1.9 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.3 | GO:0002446 | neutrophil mediated immunity(GO:0002446) |
| 0.0 | 3.1 | GO:0044042 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 1.6 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.3 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.1 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 2.1 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 1.8 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.1 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.0 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.0 | 0.2 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 1.0 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.2 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.6 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.6 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 1.2 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 1.4 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 1.8 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.8 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.8 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.6 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 1.1 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.7 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.0 | 0.9 | GO:1903078 | positive regulation of protein localization to plasma membrane(GO:1903078) |
| 0.0 | 0.8 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.3 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 3.7 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 0.2 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 0.3 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 3.9 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.4 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.0 | 2.1 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.0 | 1.5 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.1 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.0 | 0.3 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 2.6 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 1.1 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.8 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 4.8 | GO:0006631 | fatty acid metabolic process(GO:0006631) |
| 0.0 | 0.9 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.0 | 1.6 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 2.4 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 2.0 | GO:0007179 | transforming growth factor beta receptor signaling pathway(GO:0007179) |
| 0.0 | 3.9 | GO:0050890 | cognition(GO:0050890) |
| 0.0 | 0.3 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.0 | 0.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.2 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.7 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 1.1 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 1.9 | GO:0035304 | regulation of protein dephosphorylation(GO:0035304) |
| 0.0 | 0.3 | GO:0045187 | circadian sleep/wake cycle process(GO:0022410) regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) circadian sleep/wake cycle, sleep(GO:0050802) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.4 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 46.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 3.0 | 29.8 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 2.4 | 7.1 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 1.3 | 10.0 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 1.2 | 3.5 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 1.1 | 3.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 1.1 | 6.4 | GO:0032437 | cuticular plate(GO:0032437) |
| 1.0 | 5.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.9 | 0.9 | GO:0098803 | respiratory chain complex(GO:0098803) |
| 0.8 | 7.0 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.8 | 3.0 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.7 | 8.5 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.7 | 2.7 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.7 | 2.0 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.7 | 3.4 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.7 | 5.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.6 | 5.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.5 | 2.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.5 | 5.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.4 | 1.7 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.4 | 1.7 | GO:0008623 | CHRAC(GO:0008623) |
| 0.4 | 2.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.4 | 2.6 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.4 | 3.9 | GO:0097433 | dense body(GO:0097433) |
| 0.3 | 2.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.3 | 2.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.3 | 1.7 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.3 | 1.3 | GO:0032280 | axonemal microtubule(GO:0005879) symmetric synapse(GO:0032280) |
| 0.3 | 4.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.3 | 5.0 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.3 | 5.1 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.3 | 0.9 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.3 | 16.4 | GO:0002102 | podosome(GO:0002102) |
| 0.3 | 1.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.3 | 0.8 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.3 | 3.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 2.7 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.2 | 5.1 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.2 | 1.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.2 | 1.4 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.2 | 6.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 1.6 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.2 | 1.1 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.2 | 1.7 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.2 | 3.9 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.2 | 2.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.2 | 5.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.2 | 2.8 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 5.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 1.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 1.0 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 2.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.2 | 2.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.2 | 0.8 | GO:1990462 | omegasome(GO:1990462) |
| 0.2 | 1.9 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 1.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 3.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 1.6 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.2 | 5.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 1.2 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.2 | 4.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 2.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.2 | 2.6 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 0.2 | 5.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 0.9 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 1.8 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 4.1 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.6 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 2.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 4.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.5 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 2.8 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 6.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.9 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 0.5 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 1.7 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 11.1 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 2.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.7 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 5.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 2.0 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 1.8 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 1.0 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 2.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 2.1 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.8 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 4.5 | GO:0000791 | euchromatin(GO:0000791) |
| 0.1 | 5.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 6.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 4.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 0.3 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 3.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.9 | GO:0036452 | ESCRT complex(GO:0036452) |
| 0.1 | 1.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.3 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.1 | 5.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 1.8 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 0.7 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.8 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 5.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 2.9 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 3.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 5.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.3 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.8 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 1.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 1.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 2.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.5 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 3.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 7.8 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.2 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 6.5 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 19.8 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 0.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 14.1 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 2.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 1.2 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 1.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 4.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.6 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.5 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 2.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.8 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 2.0 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 22.0 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.3 | GO:0032279 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 3.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 9.9 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 2.3 | 7.0 | GO:0016034 | maleylacetoacetate isomerase activity(GO:0016034) |
| 2.2 | 13.3 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 2.0 | 6.0 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 1.7 | 1.7 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 1.7 | 6.8 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 1.7 | 8.5 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 1.7 | 10.0 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 1.4 | 4.3 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 1.3 | 4.0 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 1.2 | 3.6 | GO:0098973 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 1.2 | 7.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 1.1 | 9.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 1.1 | 6.8 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 1.1 | 4.4 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 1.1 | 3.2 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 1.1 | 5.4 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 1.0 | 8.3 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 1.0 | 5.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 1.0 | 16.3 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.9 | 5.2 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.8 | 2.4 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.8 | 3.1 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.8 | 3.1 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.8 | 31.9 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.8 | 3.0 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.7 | 2.2 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.7 | 4.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.7 | 3.5 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.7 | 4.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.7 | 5.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.6 | 2.6 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.6 | 3.9 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.5 | 2.2 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.5 | 2.2 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.5 | 2.2 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.5 | 9.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.5 | 4.0 | GO:0005534 | galactose binding(GO:0005534) |
| 0.5 | 1.4 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.5 | 1.4 | GO:0038052 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.5 | 3.4 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.4 | 2.6 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.4 | 2.6 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.4 | 1.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.4 | 11.8 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.4 | 8.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.4 | 1.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.4 | 3.8 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.4 | 1.8 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.4 | 1.8 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.4 | 4.6 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.4 | 6.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.4 | 3.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.3 | 2.7 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.3 | 13.7 | GO:0016645 | oxidoreductase activity, acting on the CH-NH group of donors(GO:0016645) |
| 0.3 | 2.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.3 | 1.6 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.3 | 4.4 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.3 | 0.9 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.3 | 7.0 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.3 | 7.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.3 | 8.5 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.3 | 0.8 | GO:0004134 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.3 | 5.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 8.1 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.2 | 3.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.2 | 2.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 0.9 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.2 | 1.6 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.2 | 2.6 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.2 | 1.7 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.2 | 2.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.2 | 1.4 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.2 | 4.1 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.2 | 3.4 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.2 | 0.7 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.2 | 1.6 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 6.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 1.9 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.2 | 4.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 1.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 0.9 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.2 | 1.9 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.2 | 2.5 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.2 | 1.5 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.2 | 6.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.6 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.1 | 1.5 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 2.6 | GO:0004559 | alpha-mannosidase activity(GO:0004559) mannosidase activity(GO:0015923) |
| 0.1 | 0.8 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 7.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 2.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 5.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 1.4 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 2.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 1.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 5.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 11.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 2.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 3.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 1.8 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.4 | GO:0001003 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.1 | 2.0 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 2.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.5 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 4.6 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 1.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 1.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.3 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.1 | 13.1 | GO:0020037 | heme binding(GO:0020037) |
| 0.1 | 0.6 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 1.0 | GO:0008061 | chitin binding(GO:0008061) |
| 0.1 | 1.6 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 1.0 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.4 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 2.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.9 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.6 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 0.8 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 3.2 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 11.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 0.7 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 2.0 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 2.6 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.5 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 1.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 2.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 0.7 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 1.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.9 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.3 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.1 | 4.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 2.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 2.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.8 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 1.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 1.0 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 1.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 5.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 10.1 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.1 | 0.6 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.0 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 7.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.8 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 1.3 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 2.0 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 6.4 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.9 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 22.4 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 3.1 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 1.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.9 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 7.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 2.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.7 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 0.1 | GO:0035870 | dITP diphosphatase activity(GO:0035870) |
| 0.0 | 0.5 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 1.2 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.4 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0048038 | quinone binding(GO:0048038) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 13.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.5 | 36.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.3 | 16.5 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.2 | 6.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 8.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 11.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 1.7 | PID EPO PATHWAY | EPO signaling pathway |
| 0.2 | 3.5 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.2 | 4.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.2 | 2.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 2.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 2.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 2.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 4.0 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 9.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 7.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 2.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.8 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 3.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 1.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 4.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 2.5 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 3.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 0.7 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 5.6 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 2.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 3.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.8 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 2.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 2.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.3 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.2 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.3 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 16.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.9 | 12.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.8 | 10.7 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.7 | 2.6 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.6 | 5.7 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.5 | 7.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.4 | 5.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.4 | 6.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.4 | 4.6 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.4 | 3.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.3 | 9.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.3 | 11.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.3 | 9.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.3 | 3.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.3 | 8.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 3.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 2.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 17.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 1.6 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.2 | 5.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 6.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 2.9 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.2 | 8.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 3.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 2.8 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 3.6 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 2.5 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 3.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 3.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 2.8 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 2.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 0.9 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 4.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 2.6 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 4.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.7 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 5.4 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 4.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 16.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 3.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 5.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 5.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 3.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.2 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.9 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.7 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |
| 0.0 | 1.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.9 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.3 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.8 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.2 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |