Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
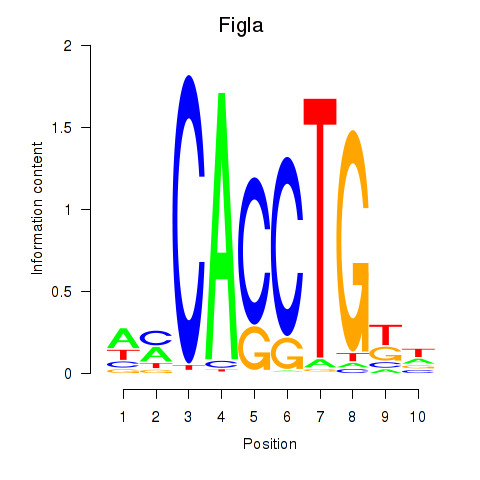
Results for Figla
Z-value: 0.72
Transcription factors associated with Figla
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Figla
|
ENSMUSG00000030001.5 | folliculogenesis specific basic helix-loop-helix |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Figla | mm39_v1_chr6_+_85994173_85994214 | 0.13 | 4.6e-01 | Click! |
Activity profile of Figla motif
Sorted Z-values of Figla motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_137157824 | 7.30 |
ENSMUST00000102522.5
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr3_-_20329823 | 7.19 |
ENSMUST00000011607.6
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr2_-_28453374 | 5.88 |
ENSMUST00000028161.6
|
Cel
|
carboxyl ester lipase |
| chr4_-_137137088 | 5.15 |
ENSMUST00000024200.7
|
Cela3a
|
chymotrypsin-like elastase family, member 3A |
| chr6_-_41012435 | 3.94 |
ENSMUST00000031931.6
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr17_-_26417982 | 3.84 |
ENSMUST00000142410.2
ENSMUST00000120333.8 ENSMUST00000039113.14 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr19_+_52252735 | 3.44 |
ENSMUST00000039652.6
|
Ins1
|
insulin I |
| chr7_-_142233270 | 3.26 |
ENSMUST00000162317.2
ENSMUST00000125933.2 ENSMUST00000105931.8 ENSMUST00000105930.8 ENSMUST00000105933.8 ENSMUST00000105932.2 ENSMUST00000000220.3 |
Ins2
|
insulin II |
| chr5_+_123214332 | 2.77 |
ENSMUST00000067505.15
ENSMUST00000111619.10 ENSMUST00000160344.2 |
Tmem120b
|
transmembrane protein 120B |
| chr14_-_20319242 | 2.44 |
ENSMUST00000024155.9
|
Kcnk16
|
potassium channel, subfamily K, member 16 |
| chr6_+_29694181 | 2.32 |
ENSMUST00000046750.14
ENSMUST00000115250.4 |
Tspan33
|
tetraspanin 33 |
| chr2_+_164611812 | 1.85 |
ENSMUST00000088248.13
ENSMUST00000001439.7 |
Ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr3_-_75177378 | 1.84 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr9_-_44253588 | 1.71 |
ENSMUST00000215091.2
|
Hmbs
|
hydroxymethylbilane synthase |
| chr9_+_107852733 | 1.54 |
ENSMUST00000035216.11
|
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr11_+_101207743 | 1.36 |
ENSMUST00000151385.2
|
Psme3
|
proteaseome (prosome, macropain) activator subunit 3 (PA28 gamma, Ki) |
| chr2_-_62313981 | 1.28 |
ENSMUST00000136686.2
ENSMUST00000102733.10 |
Gcg
|
glucagon |
| chr7_+_78563964 | 1.28 |
ENSMUST00000120331.4
|
Isg20
|
interferon-stimulated protein |
| chr12_+_109425769 | 1.18 |
ENSMUST00000173812.2
|
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr3_+_95496270 | 1.13 |
ENSMUST00000176674.8
ENSMUST00000177389.8 ENSMUST00000176755.8 ENSMUST00000177399.2 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr7_+_78564062 | 1.09 |
ENSMUST00000205981.2
|
Isg20
|
interferon-stimulated protein |
| chr3_+_95496239 | 1.08 |
ENSMUST00000177390.8
ENSMUST00000060323.12 ENSMUST00000098861.11 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr9_-_44253630 | 1.08 |
ENSMUST00000097558.5
|
Hmbs
|
hydroxymethylbilane synthase |
| chr11_-_69496655 | 1.06 |
ENSMUST00000047889.13
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide |
| chr5_-_138169509 | 1.05 |
ENSMUST00000153867.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr11_+_101207021 | 1.01 |
ENSMUST00000142640.8
ENSMUST00000019470.14 |
Psme3
|
proteaseome (prosome, macropain) activator subunit 3 (PA28 gamma, Ki) |
| chr11_+_32250069 | 0.89 |
ENSMUST00000020535.2
|
Hbq1a
|
hemoglobin, theta 1A |
| chr9_-_107960528 | 0.87 |
ENSMUST00000159372.3
ENSMUST00000160249.8 |
Rnf123
|
ring finger protein 123 |
| chr13_-_100922910 | 0.83 |
ENSMUST00000174038.2
ENSMUST00000091295.14 ENSMUST00000072119.15 |
Ccnb1
|
cyclin B1 |
| chr4_-_133600308 | 0.77 |
ENSMUST00000137486.3
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr4_-_133599616 | 0.75 |
ENSMUST00000157067.9
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr5_+_123887759 | 0.74 |
ENSMUST00000031366.12
|
Kntc1
|
kinetochore associated 1 |
| chr5_-_138169476 | 0.72 |
ENSMUST00000147920.2
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr5_-_138170077 | 0.68 |
ENSMUST00000155902.8
ENSMUST00000148879.8 |
Mcm7
|
minichromosome maintenance complex component 7 |
| chr1_-_75482975 | 0.67 |
ENSMUST00000113567.10
ENSMUST00000113565.3 |
Obsl1
|
obscurin-like 1 |
| chr6_+_40941688 | 0.66 |
ENSMUST00000076638.7
|
1810009J06Rik
|
RIKEN cDNA 1810009J06 gene |
| chr18_-_73948501 | 0.64 |
ENSMUST00000025439.5
|
Me2
|
malic enzyme 2, NAD(+)-dependent, mitochondrial |
| chr7_+_44866635 | 0.62 |
ENSMUST00000097216.5
ENSMUST00000209343.2 ENSMUST00000209678.2 |
Tead2
|
TEA domain family member 2 |
| chr14_+_70694887 | 0.60 |
ENSMUST00000003561.10
|
Phyhip
|
phytanoyl-CoA hydroxylase interacting protein |
| chrX_-_36253309 | 0.60 |
ENSMUST00000060474.14
ENSMUST00000053456.11 ENSMUST00000115239.10 |
Septin6
|
septin 6 |
| chr4_-_116228921 | 0.58 |
ENSMUST00000239239.2
ENSMUST00000239177.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr17_+_47922497 | 0.54 |
ENSMUST00000024778.3
|
Med20
|
mediator complex subunit 20 |
| chr5_+_137627431 | 0.51 |
ENSMUST00000176667.8
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr3_+_88523730 | 0.46 |
ENSMUST00000175779.8
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr7_-_24919247 | 0.46 |
ENSMUST00000058702.7
|
Dedd2
|
death effector domain-containing DNA binding protein 2 |
| chr7_+_27770655 | 0.46 |
ENSMUST00000138392.8
ENSMUST00000076648.8 |
Fcgbp
|
Fc fragment of IgG binding protein |
| chr11_+_87628356 | 0.46 |
ENSMUST00000093955.12
|
Supt4a
|
SPT4A, DSIF elongation factor subunit |
| chr7_+_24982206 | 0.46 |
ENSMUST00000165239.3
|
Cic
|
capicua transcriptional repressor |
| chr17_-_47922374 | 0.42 |
ENSMUST00000024783.9
|
Bysl
|
bystin-like |
| chr12_-_40087393 | 0.40 |
ENSMUST00000146905.2
|
Arl4a
|
ADP-ribosylation factor-like 4A |
| chr15_-_81614063 | 0.39 |
ENSMUST00000171115.3
ENSMUST00000170134.9 ENSMUST00000052374.13 |
Rangap1
|
RAN GTPase activating protein 1 |
| chr2_+_122479770 | 0.38 |
ENSMUST00000047498.15
ENSMUST00000110512.4 |
AA467197
|
expressed sequence AA467197 |
| chr9_+_108820846 | 0.37 |
ENSMUST00000198140.5
ENSMUST00000051873.15 |
Pfkfb4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr8_+_70953766 | 0.36 |
ENSMUST00000127983.2
|
Crlf1
|
cytokine receptor-like factor 1 |
| chr15_-_78657640 | 0.36 |
ENSMUST00000018313.6
|
Mfng
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr1_-_182929025 | 0.35 |
ENSMUST00000171366.7
|
Disp1
|
dispatched RND transporter family member 1 |
| chr2_-_168607166 | 0.34 |
ENSMUST00000137536.2
|
Sall4
|
spalt like transcription factor 4 |
| chr11_+_96820220 | 0.34 |
ENSMUST00000062172.6
|
Prr15l
|
proline rich 15-like |
| chr7_-_24771717 | 0.33 |
ENSMUST00000003468.10
|
Grik5
|
glutamate receptor, ionotropic, kainate 5 (gamma 2) |
| chr7_+_27259895 | 0.33 |
ENSMUST00000187032.2
|
2310022A10Rik
|
RIKEN cDNA 2310022A10 gene |
| chr7_-_29204812 | 0.32 |
ENSMUST00000183096.8
ENSMUST00000085809.11 |
Sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chrX_-_73325318 | 0.31 |
ENSMUST00000239458.2
ENSMUST00000019232.10 |
Dnase1l1
|
deoxyribonuclease 1-like 1 |
| chr5_+_122348140 | 0.31 |
ENSMUST00000196187.5
ENSMUST00000100747.3 |
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr11_+_96820091 | 0.31 |
ENSMUST00000054311.6
ENSMUST00000107636.4 |
Prr15l
|
proline rich 15-like |
| chr10_+_12936248 | 0.31 |
ENSMUST00000193426.6
|
Plagl1
|
pleiomorphic adenoma gene-like 1 |
| chr11_+_63019799 | 0.31 |
ENSMUST00000108702.8
|
Pmp22
|
peripheral myelin protein 22 |
| chr2_-_170248421 | 0.30 |
ENSMUST00000154650.8
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr17_-_88105422 | 0.30 |
ENSMUST00000055221.9
|
Kcnk12
|
potassium channel, subfamily K, member 12 |
| chr4_+_130643260 | 0.30 |
ENSMUST00000030316.7
|
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr1_-_192880260 | 0.29 |
ENSMUST00000161367.2
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chrX_-_73325204 | 0.29 |
ENSMUST00000114189.10
ENSMUST00000119361.4 |
Dnase1l1
|
deoxyribonuclease 1-like 1 |
| chr15_-_89007290 | 0.28 |
ENSMUST00000109353.9
|
Tubgcp6
|
tubulin, gamma complex associated protein 6 |
| chr13_+_21364330 | 0.28 |
ENSMUST00000223065.2
|
Trim27
|
tripartite motif-containing 27 |
| chr5_+_91665474 | 0.28 |
ENSMUST00000040576.10
|
Parm1
|
prostate androgen-regulated mucin-like protein 1 |
| chr5_+_31855009 | 0.27 |
ENSMUST00000201352.4
ENSMUST00000202815.4 |
Babam2
|
BRISC and BRCA1 A complex member 2 |
| chr7_-_126817639 | 0.27 |
ENSMUST00000152267.8
ENSMUST00000106314.8 |
Septin1
|
septin 1 |
| chr5_+_122347912 | 0.27 |
ENSMUST00000143560.8
|
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr17_-_48235325 | 0.26 |
ENSMUST00000113263.8
ENSMUST00000097311.9 |
Foxp4
|
forkhead box P4 |
| chr15_+_102381705 | 0.26 |
ENSMUST00000229958.2
ENSMUST00000229184.2 ENSMUST00000230728.2 ENSMUST00000230114.2 |
Pcbp2
|
poly(rC) binding protein 2 |
| chr6_-_121058551 | 0.25 |
ENSMUST00000077159.8
ENSMUST00000207889.2 ENSMUST00000238957.2 ENSMUST00000238920.2 ENSMUST00000238968.2 |
Mical3
|
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
| chr2_-_122441719 | 0.25 |
ENSMUST00000028624.9
|
Gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr1_-_133352115 | 0.25 |
ENSMUST00000153799.8
|
Sox13
|
SRY (sex determining region Y)-box 13 |
| chr5_+_31855394 | 0.25 |
ENSMUST00000063813.11
ENSMUST00000071531.12 ENSMUST00000131995.7 ENSMUST00000114507.10 |
Babam2
|
BRISC and BRCA1 A complex member 2 |
| chr3_-_89905927 | 0.25 |
ENSMUST00000197725.5
ENSMUST00000197767.5 ENSMUST00000197786.5 ENSMUST00000079724.9 |
Hax1
|
HCLS1 associated X-1 |
| chr1_+_128031055 | 0.25 |
ENSMUST00000188381.7
ENSMUST00000187900.7 ENSMUST00000036288.11 |
R3hdm1
|
R3H domain containing 1 |
| chr12_-_40088024 | 0.25 |
ENSMUST00000101472.4
|
Arl4a
|
ADP-ribosylation factor-like 4A |
| chr5_+_31855304 | 0.25 |
ENSMUST00000114515.9
|
Babam2
|
BRISC and BRCA1 A complex member 2 |
| chr11_+_43419586 | 0.24 |
ENSMUST00000050574.7
|
Ccnjl
|
cyclin J-like |
| chr11_-_100036792 | 0.24 |
ENSMUST00000007317.8
|
Krt19
|
keratin 19 |
| chr16_+_49620883 | 0.24 |
ENSMUST00000229640.2
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr7_+_18828519 | 0.24 |
ENSMUST00000049454.6
|
Six5
|
sine oculis-related homeobox 5 |
| chr1_+_75358758 | 0.24 |
ENSMUST00000148515.8
ENSMUST00000113590.8 |
Speg
|
SPEG complex locus |
| chr12_+_113061819 | 0.24 |
ENSMUST00000109727.9
ENSMUST00000009099.13 ENSMUST00000109723.8 ENSMUST00000109726.8 ENSMUST00000069690.5 |
Mta1
|
metastasis associated 1 |
| chr6_-_8778439 | 0.24 |
ENSMUST00000115520.8
ENSMUST00000038403.12 ENSMUST00000115518.8 |
Ica1
|
islet cell autoantigen 1 |
| chr6_-_8778106 | 0.23 |
ENSMUST00000151758.2
ENSMUST00000115519.8 ENSMUST00000153390.8 |
Ica1
|
islet cell autoantigen 1 |
| chr8_+_46338498 | 0.22 |
ENSMUST00000034053.7
|
Pdlim3
|
PDZ and LIM domain 3 |
| chr4_-_135221926 | 0.22 |
ENSMUST00000102549.10
|
Nipal3
|
NIPA-like domain containing 3 |
| chr4_-_134431534 | 0.21 |
ENSMUST00000054096.13
ENSMUST00000038628.4 |
Man1c1
|
mannosidase, alpha, class 1C, member 1 |
| chr11_-_97171258 | 0.21 |
ENSMUST00000165216.8
|
Npepps
|
aminopeptidase puromycin sensitive |
| chr11_-_82761954 | 0.21 |
ENSMUST00000108173.10
ENSMUST00000071152.14 |
Rffl
|
ring finger and FYVE like domain containing protein |
| chr5_-_73349191 | 0.20 |
ENSMUST00000176910.3
|
Fryl
|
FRY like transcription coactivator |
| chr7_+_48993197 | 0.20 |
ENSMUST00000207743.2
|
Nav2
|
neuron navigator 2 |
| chr11_-_97171294 | 0.20 |
ENSMUST00000167806.8
ENSMUST00000172108.8 ENSMUST00000001480.14 |
Npepps
|
aminopeptidase puromycin sensitive |
| chr3_-_89905547 | 0.19 |
ENSMUST00000199740.2
ENSMUST00000198782.2 |
Hax1
|
HCLS1 associated X-1 |
| chr6_-_12749192 | 0.19 |
ENSMUST00000172356.8
|
Thsd7a
|
thrombospondin, type I, domain containing 7A |
| chr15_-_51728893 | 0.19 |
ENSMUST00000022925.10
|
Eif3h
|
eukaryotic translation initiation factor 3, subunit H |
| chr7_-_126817475 | 0.18 |
ENSMUST00000106313.8
ENSMUST00000142356.3 |
Septin1
|
septin 1 |
| chr11_-_97171185 | 0.18 |
ENSMUST00000168743.8
|
Npepps
|
aminopeptidase puromycin sensitive |
| chr9_-_20809888 | 0.18 |
ENSMUST00000004206.10
|
Eif3g
|
eukaryotic translation initiation factor 3, subunit G |
| chr9_+_50679528 | 0.18 |
ENSMUST00000042391.13
|
Fdxacb1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr11_-_54853729 | 0.18 |
ENSMUST00000108885.8
ENSMUST00000102730.9 ENSMUST00000018482.13 ENSMUST00000108886.8 ENSMUST00000102731.8 |
Tnip1
|
TNFAIP3 interacting protein 1 |
| chr15_-_101268036 | 0.18 |
ENSMUST00000077196.6
|
Krt80
|
keratin 80 |
| chr16_+_17269845 | 0.17 |
ENSMUST00000006293.5
ENSMUST00000231228.2 |
Crkl
|
v-crk avian sarcoma virus CT10 oncogene homolog-like |
| chr11_+_72332167 | 0.17 |
ENSMUST00000045633.6
|
Mybbp1a
|
MYB binding protein (P160) 1a |
| chr11_+_71640739 | 0.17 |
ENSMUST00000150531.2
|
Wscd1
|
WSC domain containing 1 |
| chr11_+_104468107 | 0.17 |
ENSMUST00000106956.10
|
Myl4
|
myosin, light polypeptide 4 |
| chr2_+_112096154 | 0.16 |
ENSMUST00000110991.9
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr17_+_35413415 | 0.16 |
ENSMUST00000025262.6
ENSMUST00000173600.2 |
Ltb
|
lymphotoxin B |
| chr10_-_80492315 | 0.16 |
ENSMUST00000126980.8
|
Btbd2
|
BTB (POZ) domain containing 2 |
| chr14_-_70415117 | 0.16 |
ENSMUST00000022681.11
|
Pdlim2
|
PDZ and LIM domain 2 |
| chr17_+_24692858 | 0.15 |
ENSMUST00000054946.10
ENSMUST00000238986.2 ENSMUST00000164508.2 |
Bricd5
|
BRICHOS domain containing 5 |
| chr7_-_131012051 | 0.15 |
ENSMUST00000154602.8
ENSMUST00000142349.3 |
Ikzf5
|
IKAROS family zinc finger 5 |
| chr11_-_76514334 | 0.15 |
ENSMUST00000238684.2
|
Abr
|
active BCR-related gene |
| chr4_+_132001680 | 0.15 |
ENSMUST00000030731.11
ENSMUST00000105963.2 |
Taf12
|
TATA-box binding protein associated factor 12 |
| chrX_-_103714653 | 0.15 |
ENSMUST00000042070.6
|
Zdhhc15
|
zinc finger, DHHC domain containing 15 |
| chr2_+_32041083 | 0.15 |
ENSMUST00000036691.14
ENSMUST00000069817.15 |
Prrc2b
|
proline-rich coiled-coil 2B |
| chr17_-_43813664 | 0.14 |
ENSMUST00000024707.9
ENSMUST00000117137.8 |
Mep1a
|
meprin 1 alpha |
| chr9_-_30833748 | 0.14 |
ENSMUST00000065112.7
|
Adamts15
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 15 |
| chr11_+_104467791 | 0.13 |
ENSMUST00000106957.8
|
Myl4
|
myosin, light polypeptide 4 |
| chr5_+_20112704 | 0.13 |
ENSMUST00000115267.7
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr1_-_52271455 | 0.13 |
ENSMUST00000114512.8
|
Gls
|
glutaminase |
| chr10_-_21036824 | 0.13 |
ENSMUST00000020158.9
|
Myb
|
myeloblastosis oncogene |
| chr1_+_128069677 | 0.13 |
ENSMUST00000187023.7
|
R3hdm1
|
R3H domain containing 1 |
| chr8_-_70892204 | 0.13 |
ENSMUST00000076615.6
|
Crtc1
|
CREB regulated transcription coactivator 1 |
| chr2_+_14878480 | 0.13 |
ENSMUST00000114719.7
|
Cacnb2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr4_+_129948803 | 0.12 |
ENSMUST00000123617.8
|
Col16a1
|
collagen, type XVI, alpha 1 |
| chr1_+_128069716 | 0.12 |
ENSMUST00000187557.2
|
R3hdm1
|
R3H domain containing 1 |
| chr10_-_21036792 | 0.12 |
ENSMUST00000188495.8
|
Myb
|
myeloblastosis oncogene |
| chr11_+_70323452 | 0.12 |
ENSMUST00000084954.13
ENSMUST00000108568.10 ENSMUST00000079056.9 ENSMUST00000102564.11 ENSMUST00000124943.8 ENSMUST00000150076.8 ENSMUST00000102563.2 |
Arrb2
|
arrestin, beta 2 |
| chr1_-_128030148 | 0.12 |
ENSMUST00000086614.12
|
Zranb3
|
zinc finger, RAN-binding domain containing 3 |
| chr17_-_29566774 | 0.12 |
ENSMUST00000095427.12
ENSMUST00000118366.9 |
Mtch1
|
mitochondrial carrier 1 |
| chr8_+_72889073 | 0.12 |
ENSMUST00000003575.11
|
Tpm4
|
tropomyosin 4 |
| chr10_-_126866682 | 0.11 |
ENSMUST00000040560.11
|
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr2_-_34716083 | 0.11 |
ENSMUST00000113077.8
ENSMUST00000028220.10 |
Fbxw2
|
F-box and WD-40 domain protein 2 |
| chr6_-_135224881 | 0.11 |
ENSMUST00000087729.12
|
Gsg1
|
germ cell associated 1 |
| chr6_-_12749409 | 0.11 |
ENSMUST00000119581.7
|
Thsd7a
|
thrombospondin, type I, domain containing 7A |
| chr17_-_36011378 | 0.11 |
ENSMUST00000119825.8
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr18_+_76192529 | 0.11 |
ENSMUST00000167921.2
|
Zbtb7c
|
zinc finger and BTB domain containing 7C |
| chr4_-_135221810 | 0.10 |
ENSMUST00000105856.9
|
Nipal3
|
NIPA-like domain containing 3 |
| chr19_+_56536685 | 0.10 |
ENSMUST00000071423.7
|
Nhlrc2
|
NHL repeat containing 2 |
| chr9_+_31191820 | 0.10 |
ENSMUST00000117389.8
ENSMUST00000215499.2 |
Prdm10
|
PR domain containing 10 |
| chr6_+_54303837 | 0.10 |
ENSMUST00000059138.6
|
Prr15
|
proline rich 15 |
| chr17_-_32607859 | 0.10 |
ENSMUST00000087703.12
ENSMUST00000170603.3 |
Wiz
|
widely-interspaced zinc finger motifs |
| chrX_-_74460137 | 0.10 |
ENSMUST00000033542.11
|
Mtcp1
|
mature T cell proliferation 1 |
| chr9_+_64142483 | 0.09 |
ENSMUST00000039011.4
|
Uchl4
|
ubiquitin carboxyl-terminal esterase L4 |
| chr10_-_75768302 | 0.09 |
ENSMUST00000120281.8
ENSMUST00000000924.13 |
Mmp11
|
matrix metallopeptidase 11 |
| chr1_+_75351914 | 0.09 |
ENSMUST00000087122.12
|
Speg
|
SPEG complex locus |
| chr1_+_156386327 | 0.08 |
ENSMUST00000173929.8
|
Abl2
|
v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene) |
| chr10_-_126866658 | 0.08 |
ENSMUST00000120547.2
ENSMUST00000152054.8 |
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr19_-_5738177 | 0.07 |
ENSMUST00000068169.12
|
Pcnx3
|
pecanex homolog 3 |
| chr9_+_57468217 | 0.07 |
ENSMUST00000045791.11
ENSMUST00000216986.2 |
Scamp2
|
secretory carrier membrane protein 2 |
| chr11_-_79394904 | 0.07 |
ENSMUST00000164465.3
|
Omg
|
oligodendrocyte myelin glycoprotein |
| chr19_-_5399368 | 0.07 |
ENSMUST00000238111.2
|
Cst6
|
cystatin E/M |
| chr15_-_63869818 | 0.07 |
ENSMUST00000164532.3
|
Cyrib
|
CYFIP related Rac1 interactor B |
| chr9_+_105535585 | 0.07 |
ENSMUST00000186943.2
|
Pik3r4
|
phosphoinositide-3-kinase regulatory subunit 4 |
| chr1_-_126665904 | 0.07 |
ENSMUST00000160693.2
|
Nckap5
|
NCK-associated protein 5 |
| chr7_+_90739904 | 0.07 |
ENSMUST00000107196.10
ENSMUST00000074273.10 |
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chrX_-_73412078 | 0.07 |
ENSMUST00000155676.8
|
Ubl4a
|
ubiquitin-like 4A |
| chr11_+_120675131 | 0.07 |
ENSMUST00000116305.8
|
Gps1
|
G protein pathway suppressor 1 |
| chr11_-_99932373 | 0.06 |
ENSMUST00000056362.3
|
Krt34
|
keratin 34 |
| chr9_+_107431776 | 0.06 |
ENSMUST00000010211.7
|
Rassf1
|
Ras association (RalGDS/AF-6) domain family member 1 |
| chr13_+_76532470 | 0.06 |
ENSMUST00000125209.8
|
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr16_-_16377982 | 0.06 |
ENSMUST00000161861.8
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr5_+_146168020 | 0.06 |
ENSMUST00000161181.8
ENSMUST00000161652.8 ENSMUST00000031640.15 ENSMUST00000159467.2 |
Cdk8
|
cyclin-dependent kinase 8 |
| chr4_-_41731142 | 0.06 |
ENSMUST00000171251.8
|
Arid3c
|
AT rich interactive domain 3C (BRIGHT-like) |
| chr2_-_92201342 | 0.05 |
ENSMUST00000176810.8
ENSMUST00000090582.11 ENSMUST00000068586.13 |
Large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr5_+_20112500 | 0.05 |
ENSMUST00000101558.10
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr14_+_54183465 | 0.05 |
ENSMUST00000197130.5
ENSMUST00000103677.3 |
Trdv2-1
|
T cell receptor delta variable 2-1 |
| chr3_+_51324022 | 0.05 |
ENSMUST00000192419.6
|
Naa15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr19_+_47167554 | 0.05 |
ENSMUST00000235290.2
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr4_-_133484080 | 0.05 |
ENSMUST00000008024.7
|
Arid1a
|
AT rich interactive domain 1A (SWI-like) |
| chr10_-_41894360 | 0.05 |
ENSMUST00000162405.8
ENSMUST00000095729.11 ENSMUST00000161081.2 ENSMUST00000160262.9 |
Armc2
|
armadillo repeat containing 2 |
| chr7_+_28092370 | 0.04 |
ENSMUST00000003529.9
|
Paf1
|
Paf1, RNA polymerase II complex component |
| chr16_-_92494203 | 0.04 |
ENSMUST00000113956.10
|
Runx1
|
runt related transcription factor 1 |
| chr3_-_64197130 | 0.04 |
ENSMUST00000233531.2
ENSMUST00000176328.9 |
Vmn2r3
|
vomeronasal 2, receptor 3 |
| chr3_-_116601451 | 0.04 |
ENSMUST00000159670.3
|
Agl
|
amylo-1,6-glucosidase, 4-alpha-glucanotransferase |
| chr2_-_58247764 | 0.04 |
ENSMUST00000112608.9
ENSMUST00000112607.3 ENSMUST00000028178.14 |
Acvr1c
|
activin A receptor, type IC |
| chr7_+_24990596 | 0.04 |
ENSMUST00000164820.2
|
Cic
|
capicua transcriptional repressor |
| chr9_+_102597486 | 0.04 |
ENSMUST00000130602.2
|
Amotl2
|
angiomotin-like 2 |
| chr8_+_66838927 | 0.04 |
ENSMUST00000039540.12
ENSMUST00000110253.3 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr7_-_28092113 | 0.04 |
ENSMUST00000003536.9
|
Med29
|
mediator complex subunit 29 |
| chr3_+_95341698 | 0.04 |
ENSMUST00000102749.11
ENSMUST00000090804.12 ENSMUST00000107161.8 ENSMUST00000107160.8 ENSMUST00000015666.17 |
Arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr1_+_156386414 | 0.04 |
ENSMUST00000166172.9
ENSMUST00000027888.13 |
Abl2
|
v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene) |
| chr15_-_38300937 | 0.04 |
ENSMUST00000227920.2
ENSMUST00000074043.7 ENSMUST00000228416.2 |
Klf10
|
Kruppel-like factor 10 |
| chr11_-_99742434 | 0.03 |
ENSMUST00000107437.2
|
Krtap4-16
|
keratin associated protein 4-16 |
| chr10_-_128579879 | 0.03 |
ENSMUST00000026414.9
|
Dgka
|
diacylglycerol kinase, alpha |
| chr9_-_15023396 | 0.03 |
ENSMUST00000159985.2
|
Hephl1
|
hephaestin-like 1 |
| chr5_-_103777145 | 0.03 |
ENSMUST00000031263.2
|
Slc10a6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6 |
| chr1_-_80643024 | 0.03 |
ENSMUST00000187774.7
|
Dock10
|
dedicator of cytokinesis 10 |
| chr3_+_65435878 | 0.03 |
ENSMUST00000130705.2
|
Tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Figla
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:1990535 | negative regulation of NAD(P)H oxidase activity(GO:0033861) neuron projection maintenance(GO:1990535) |
| 0.9 | 2.8 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.4 | 2.4 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.4 | 5.9 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.3 | 1.9 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.2 | 1.1 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.2 | 0.8 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.2 | 2.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 1.5 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.2 | 1.5 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.2 | 1.3 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 2.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 2.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 0.3 | GO:0035638 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.1 | 3.8 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.1 | 0.3 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.1 | 0.6 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.1 | 12.1 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.1 | 0.5 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.4 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 | 0.2 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.3 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.6 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.5 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.4 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 1.2 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.0 | 0.2 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.5 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.5 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.5 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.2 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.6 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.3 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.3 | GO:1901250 | regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.0 | 0.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.1 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.3 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.2 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.2 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.0 | 0.6 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.4 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.2 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.1 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.8 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.2 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.0 | 0.3 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 | 0.5 | GO:0043496 | regulation of protein homodimerization activity(GO:0043496) |
| 0.0 | 1.9 | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway(GO:2001237) |
| 0.0 | 0.0 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.4 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 0.7 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.2 | 0.8 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.2 | 2.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 0.6 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.2 | 2.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 5.9 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 3.3 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.1 | 1.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.4 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 0.8 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.4 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 0.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 0.2 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.1 | 1.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 2.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 2.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.3 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.3 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.4 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.5 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 27.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.3 | GO:0043218 | compact myelin(GO:0043218) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0050253 | sterol esterase activity(GO:0004771) retinyl-palmitate esterase activity(GO:0050253) |
| 0.9 | 2.8 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.5 | 2.4 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.3 | 3.8 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.3 | 1.5 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.2 | 7.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 2.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 0.6 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.8 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 2.5 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.4 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.6 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 6.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 15.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 2.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.3 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.4 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.4 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.6 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 1.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 1.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.3 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.0 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 0.0 | 0.0 | GO:0038100 | nodal binding(GO:0038100) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 13.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 3.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.5 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.5 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 2.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 2.7 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 2.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.5 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 2.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.1 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |